Database Of Space Life Science Investigations And Bioinformatics Of Microbiology In Extreme Environments
Biological experiments performed in space crafts like space stations, space shuttles, and recoverable satellites has enabled extensive spaceflight life investigations (SLIs).
In particular, SLIs have revealed distinguished space effects on microbial growth, survival, metabolite production, biofilm formation, virulence development and drug resistant mutations. These provide unique perspectives to ground-based microbiology and new opportunities for industrial pharmaceutical and metabolite productions.
SLIs are with specialized experimental setups, analysis methods and research outcomes, which can be accessed by established databases National Aeronautics and Space Administration (NASA) Life Science Data Archive, Erasmus Experiment Archive, and NASA GeneLab. The increasing research across diverse fields may be better facilitated by databases of convenient search facilities and categorized presentation of comprehensive contents.
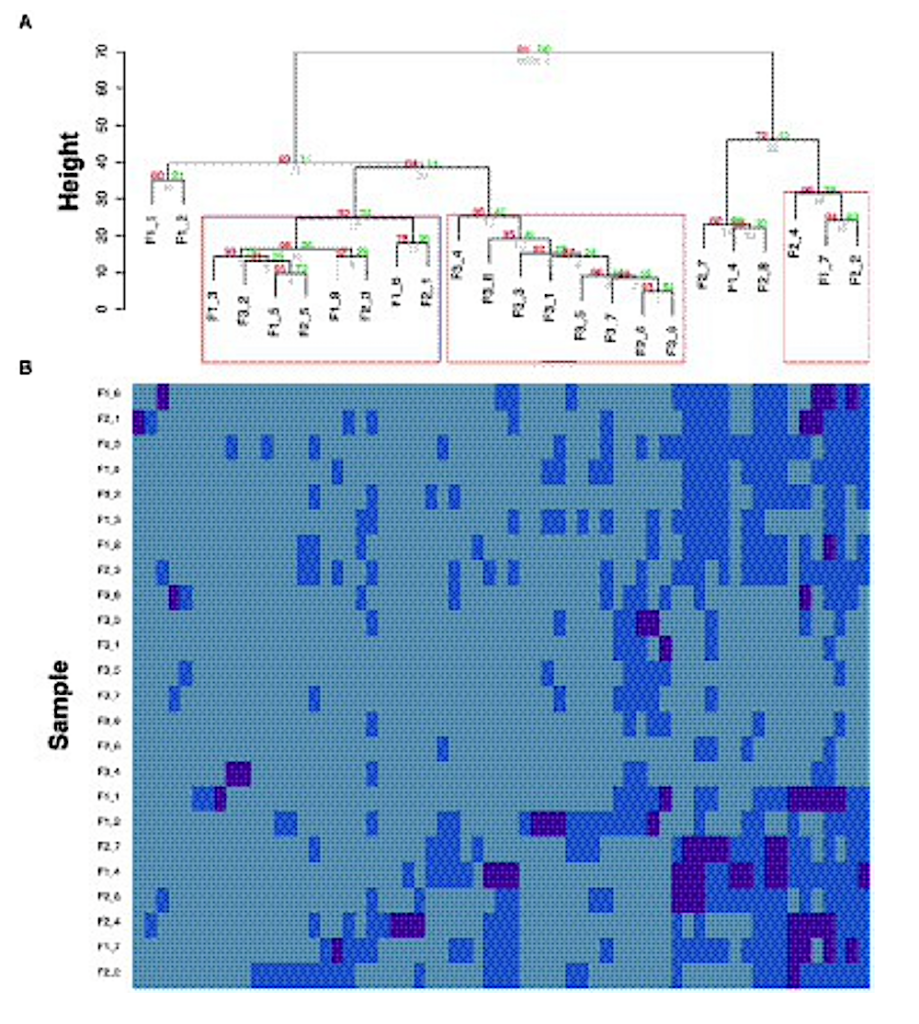
Temporal comparison of AMR-gene profiles (modified from a figure in Urbaniak et al., 2018). (A) Hierarchical clustering of the AMR-genes. Red values are AU (Approximately Unbiased) p-values, and green values are Bootstrap probability values. Clusters with AU p-values larger than 90% are highlighted by rectangles, meaning that there is a 90% certainty of these clusters being a distinct group. F“x” refers to the flight number, followed by the location. (B) Heatmap of centered log ratio transformed gene expression data. Deep blue boxes represent samples that have higher gene expression then the geometric mean expression (light blue), which has been calculated from all samples, which has been calculated from all samples.
We therefore developed the Space Life Investigation Database (SpaceLID) http://bidd.group/spacelid/, which collected SLIs from published academic papers. Currently, this database provides detailed menu search facilities and categorized contents about the studied phenomena, materials, experimental procedures, analysis methods, and research outcomes of 448 SLIs of 90 species (microbial, plant, animal, human), 81 foods and 106 pharmaceuticals, including 232 SLIs not covered by the established databases.
The potential applications of SpaceLID are illustrated by the examples of published experimental design and bioinformatic analysis of spaceflight microbial phenomena.
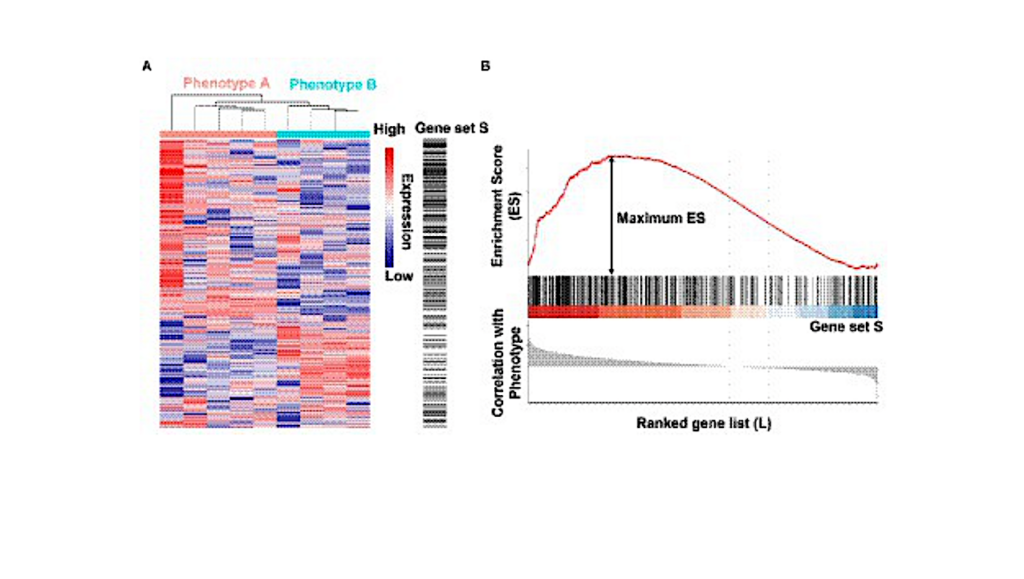
Illustration of the gene set enrichment analysis (GSEA) method. (A) A gene expression dataset sorted by correlation with phenotype, the corresponding heatmap, and the “gene tags,” i.e., location of genes from a set S within the sorted list. (B) Plot of the running sum for S in the data set, including the location of the maximum enrichment score (ES) and the leading-edge subset. The figures were drawn using data from GSE35958, only used to show the principle of GSEA method.
Junyong Wang, Tao Wang; Tao Wang, Xian ZengXian Zeng, Shanshan WangShanshan Wang, Zijie YuZijie Yu, Yiqi WeiYiqi Wei, Mengna CaiMengna Cai, Xin-Yi Chu, Yu Zong Chen, Yufen Zhao,
Database of space life investigations and bioinformatics of microbiology in extreme environments, Frontiers in Microbiology
Astrobiology