Triplet-Encoded Prebiotic RNA Aminoacylation
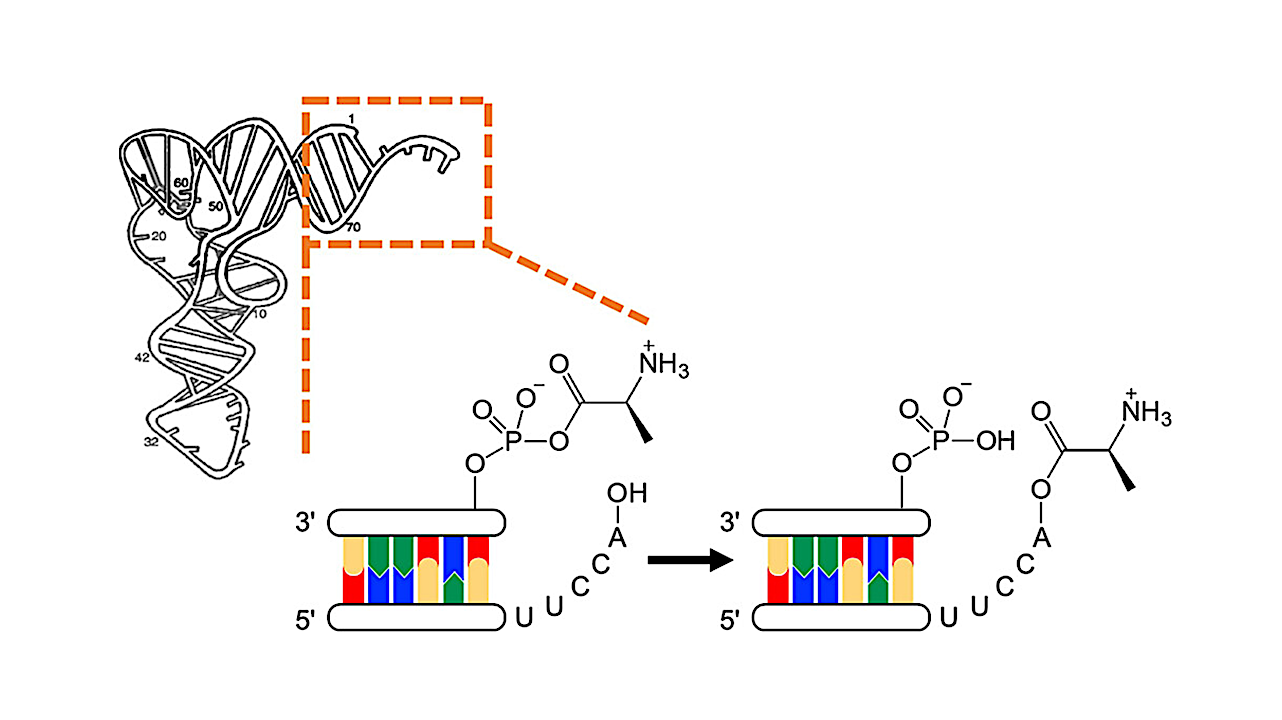
The encoding step of translation involves attachment of amino acids to cognate tRNAs by aminoacyl-tRNA synthetases, themselves the product of coded peptide synthesis. So, the question arises─before these enzymes evolved, how were primordial tRNAs selectively aminoacylated?
Here, we demonstrate enzyme-free, sequence-dependent, chemoselective aminoacylation of RNA. We investigated two potentially prebiotic routes to aminoacyl-tRNA acceptor stem-overhang mimics and analyzed those oligonucleotides undergoing the most efficient aminoacylation. Overhang sequences do not significantly influence the chemoselectivity of aminoacylation by either route.
For aminoacyl-transfer from a mixed anhydride donor strand, the chemoselectivity and stereoselectivity of aminoacylation depend on the terminal three base pairs of the stem. The results support early suggestions of a second genetic code in the acceptor stem.
https://pubs.acs.org/doi/10.1021/jacs.3c03931
J. Am. Chem. Soc. 2023, 145, 29, 15971–15980
Triplet-Encoded Prebiotic RNA Aminoacylation (open access)
Astrobiology