The Genetic Response Of An Earth Plant – Off World – In Microgravity
Spaceflight presents a multifaceted environment for plants, combining the effects on growth of many stressors and factors including altered gravity, the influence of experiment hardware, and increased radiation exposure.
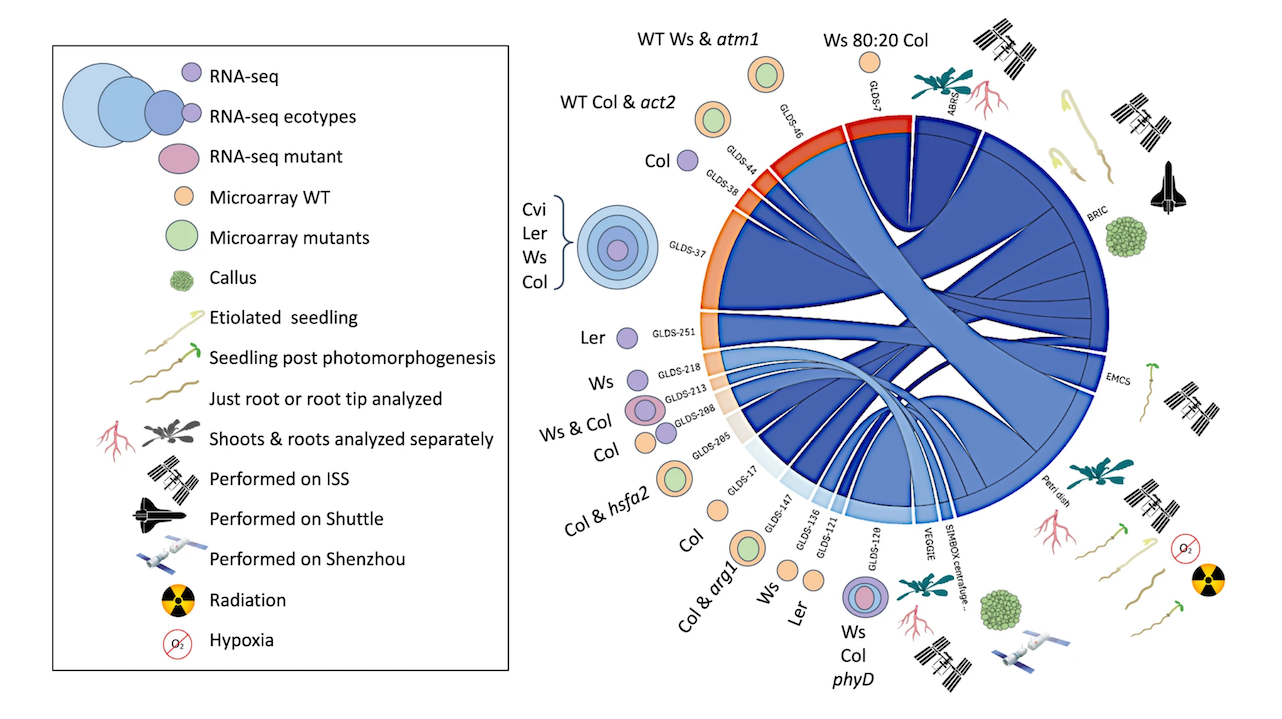
To help understand the plant response to this complex suite of factors this study compared transcriptomic analysis of 15 Arabidopsis thaliana spaceflight experiments deposited in the National Aeronautics and Space Administration’s GeneLab data repository. These data were reanalyzed for genes showing significant differential expression in spaceflight versus ground controls using a single common computational pipeline for either the microarray or the RNA-seq datasets.
Such a standardized approach to analysis should greatly increase the robustness of comparisons made between datasets. This analysis was coupled with extensive cross-referencing to a curated matrix of metadata associated with these experiments.
Our study reveals that factors such as analysis type (i.e., microarray versus RNA-seq) or environmental and hardware conditions have important confounding effects on comparisons seeking to define plant reactions to spaceflight. The metadata matrix allows selection of studies with high similarity scores, i.e., that share multiple elements of experimental design, such as plant age or flight hardware.
Comparisons between these studies then helps reduce the complexity in drawing conclusions arising from comparisons made between experiments with very different designs.
Meta-analysis of the space flight and microgravity response of the Arabidopsis plant transcriptome, Nature (open access)
Meta-analysis of the space flight and microgravity response of the Arabidopsis plant transcriptome, NPJ MIcrogravity via PubMed (open access)
Richard Barker, Colin P. S. Kruse, Christina Johnson, Amanda Saravia-Butler, Homer Fogle, Hyun-Seok Chang, Ralph Møller Trane, Noah Kinscherf, Alicia Villacampa, Aránzazu Manzano, Raúl Herranz, Laurence B. Davin, Norman G. Lewis, Imara Perera, Chris Wolverton, Parul Gupta, Pankaj Jaiswal, Sigrid S. Reinsch, Sarah Wyatt & Simon Gilroy
npj Microgravity volume 9, Article number: 21 (2023)
Astrobiology