Identifying The Last Universal Common Ancestor’s Protein Domains Resolves The Order In Which The Amino Acids Were Recruited Into The Genetic Code
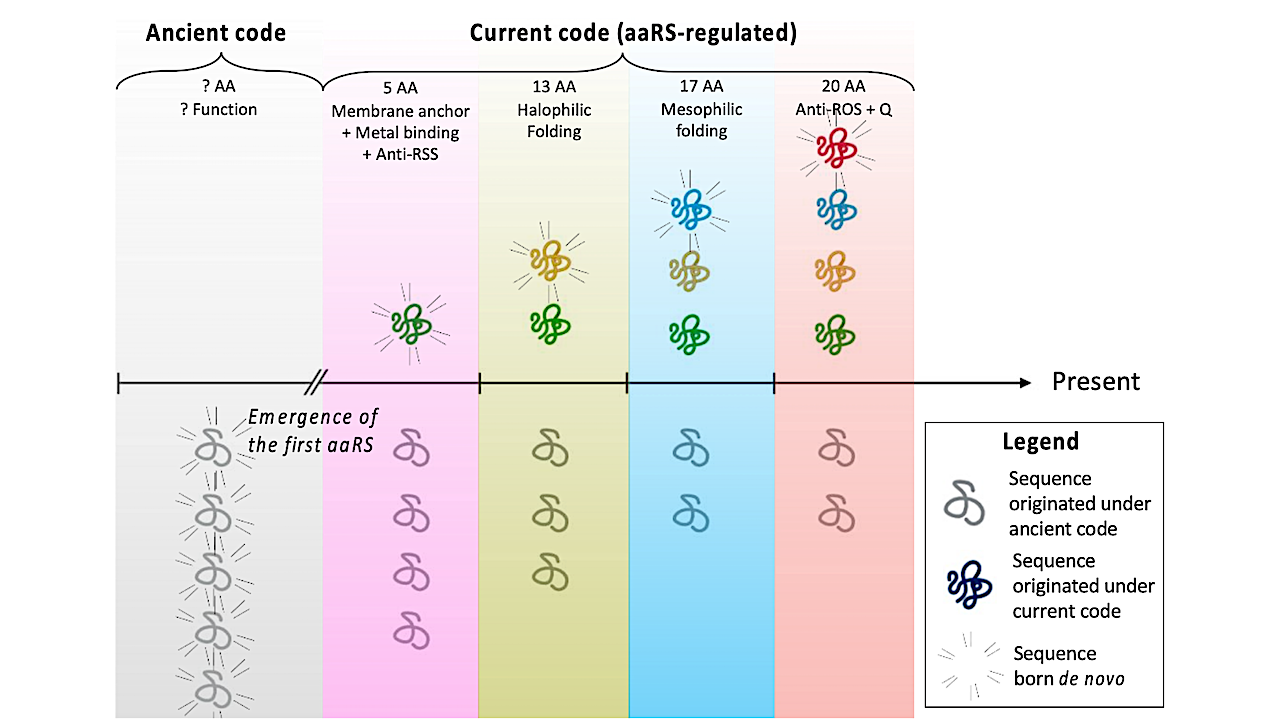
We identified protein domains that emerged early in the history of life. Protein domains whose ancestors date back to a single homolog in the Last Universal Common Ancestor (LUCA) remain depleted for amino acids believed to be added late to the genetic code.
Notable exceptions call for revisions to our understanding of the order of amino acid recruitment into the genetic code. Enrichment in ancient proteins shows that metal-binding amino acids (cysteine and histidine) and sulfur-containing amino acids (cysteine and methionine) were added much earlier than previously thought.
Sequences that had already diversified into multiple distinct copies in LUCA will tend to be even more ancient, and we therefore expected them to be more enriched for early amino acids, and depleted for late. Surprisingly, these more ancient sequences showed a different pattern, significantly less depleted for tryptophan and tyrosine, and enriched rather than depleted for phenylalanine.
This is compatible with at least some of these sequences predating the current genetic code. Their distinct enrichment patterns thus provide hints about earlier, alternative genetic codes.
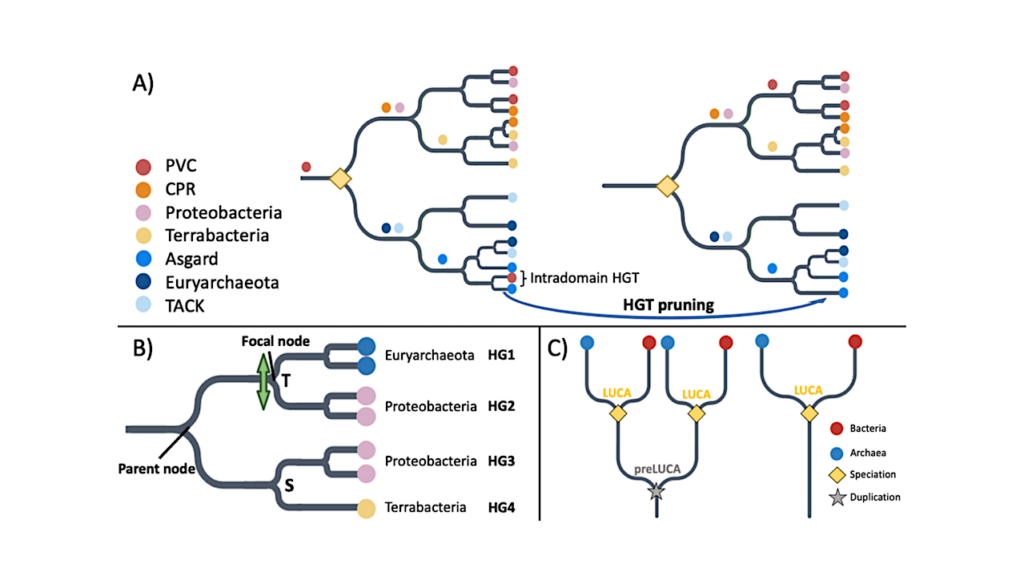
Criteria for (A) LUCA Pfam annotation, (B) Identifying HGT to be filtered, and (C) pre-LUCA Pfam annotation. A) Pruning HGT between archaea and bacteria reveals a LUCA node as dividing bacteria and archaea at the root. Colored circles are indicated just upstream of the most recent common ancestor (MRCA) of all copies of that Pfam found within the same taxonomic supergroup. We recognize five bacterial supergroups (FCB, PVC, CPR, Terrabacteria and Proteobacteria (Rinke, Schwientek et al. 2013, Brown, Hug et al. 2015)) and three archaeal supergroups (TACK, DPANN, Asgard and Euryarchaeota (Baker, De Anda et al. 2020, Shu and Huang 2021)). The yellow diamond indicates LUCA as a speciation event between archaea and bacteria. Prior to HGT pruning, PVC sequences can be found on either side of the two lineages divided by the root. After pruning intradomain HGT, four MRCAs are found one node away from the root, and 3 more MRCAs are found two nodes away from the root, fulfilling our other LUCA criterion described in the Methods, namely presence of at least three bacterial and at least two archaeal supergroup MRCAs one to two nodes away from the root. B) Criteria for pruning likely HGT between archaea and bacteria (see Methods for details). We partition into monophyletic groups of sequences in the same supergroup; in this example, there are four such groups, representing two bacterial supergroups and one archaeal supergroup. There is one ‘mixed’ node, separating an archaeal group (HG1) from a bacterial group (HG2). It is also annotated by GeneRax as a transfer ‘T’. The bacterial nature of groups 3 and 4 indicates a putative HGT direction from group 2 to group 1. Group 2 does not contain any Euryarchaeota sequences, meeting the third and final requirement for pruning of group 1. If neither Proteobacteria or Euryarchaeota sequences were present among the other descendants of the parent node, both groups 1 and 2 would be considered acceptors of a transferred Pfam and would both be pruned from the tree. C) Pre-LUCA Pfams have at least two nodes annotated as LUCA. — biorxiv.org
doi: https://doi.org/10.1101/2024.04.13.589375
Astrobiology