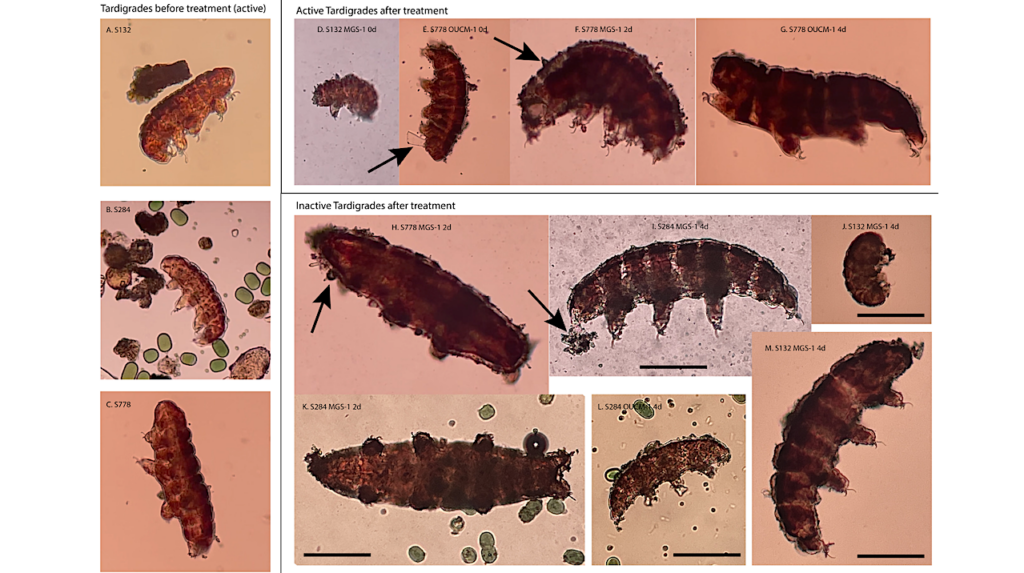
Bacteria Found On The International Space Station Are Mutating To Become Functionally Distinct
In a new scientific paper funded by an Ames Space Biology grant, Principal Investigator Dr. Kasthuri Venkateswaran of NASA’s Jet Propulsion Laboratory strains of the bacterial species Enterobacter bugandensis isolated from the International Space Station (ISS) were studied.
Thirteen strains of E. bugandensis, a bacterium notorious for being multi-drug resistant, were isolated from the ISS.
Study findings indicate under stress, the ISS isolated strains were mutated and became genetically and functionally distinct compared to their Earth counterparts.
The strains were able to viably persist in the ISS over time with a significant abundance. E. bugandensis coexisted with multiple other microorganisms, and in some cases could have helped those organisms survive.
Publication Impact: Closed human-built environments, such as the ISS, are unique areas that provide an extreme environment subject to microgravity, radiation, and elevated carbon dioxide levels. Any microorganisms introduced to these areas must adapt to thrive. By delving into microbial dynamics in extreme environments, this research opens doors to effective preventative measure for astronaut health.
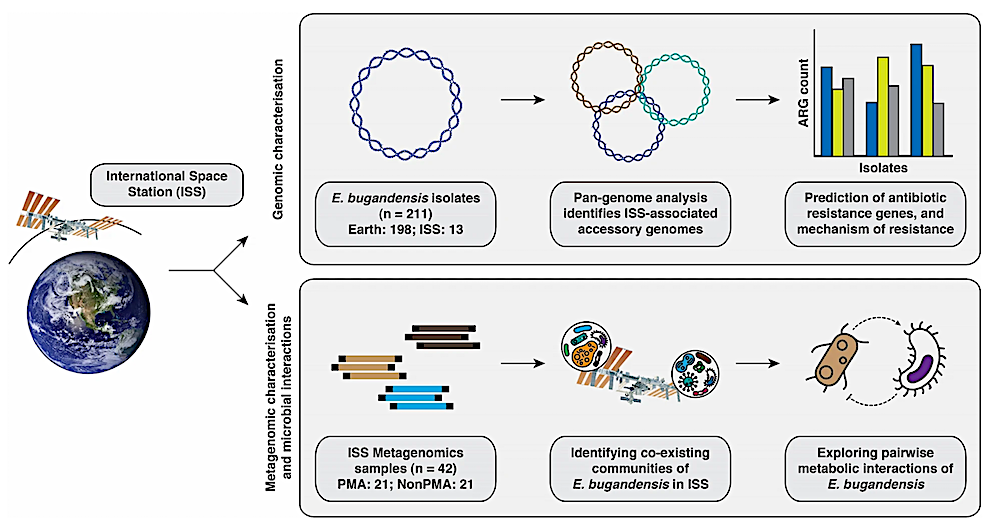
Illustrative workflow showcasing the process of comparative genomics analysis of E. bugandensis, evaluating its prevalence and metabolic interactions within the microbial community, and its successful adaptation within the ISS habitat
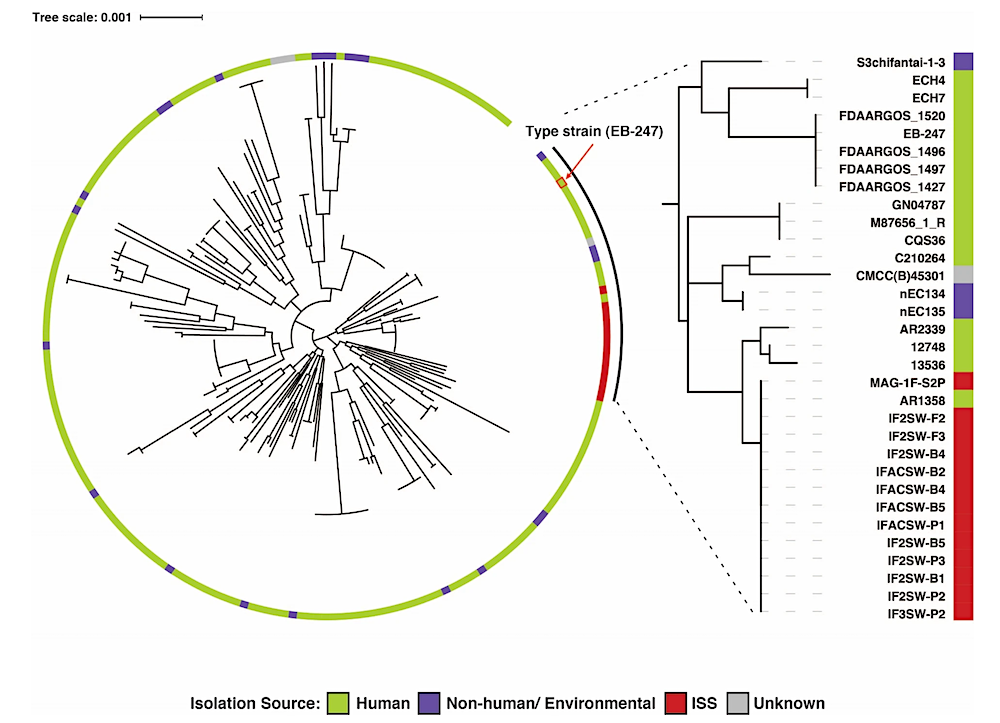
Phylogenetic tree of E. bugandensis constructed using 172 marker genes of Gammaproteobacteria showing the distinct clade of ISS isolates/MAG apart from the other Earth-based isolation sources. The clade with 32 genomes, which includes 12 ISS isolates and one MAG, is highlighted in the right-side panel
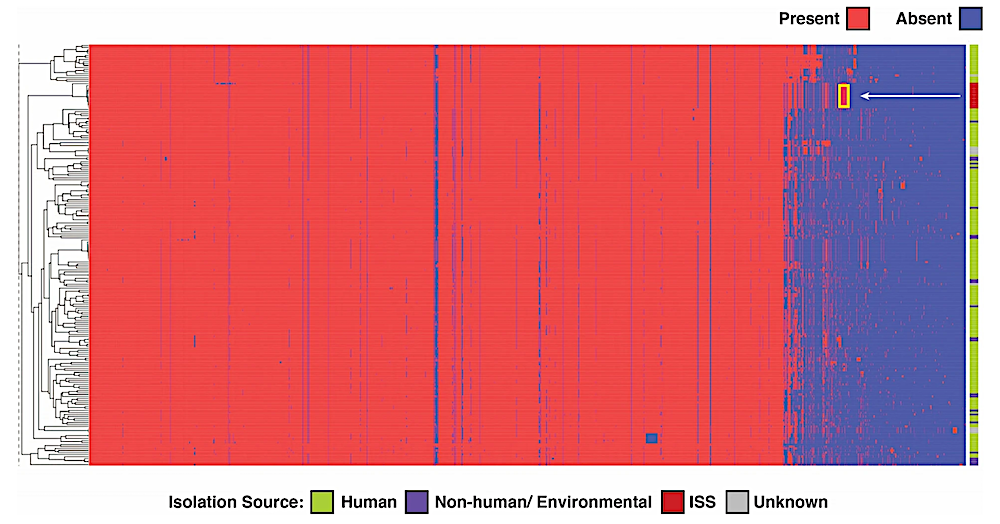
Hierarchical clustering visualisation depicting the functional profiles of E. bugandensis based on COG annotations. In this figure, columns represent COGs, and rows represent individual samples. The pointed yellow box indicates the COGs specifically present in the genomes of the ISS strains
Reference: Sengupta P, Muthamilselvi Sivabalan SK, Singh NK, Raman K, Venkateswaran K.
Genomic, functional, and metabolic enhancements in multidrug-resistant Enterobacter bugandensis facilitating its persistence and succession in the International Space Station. Microbiome. 2024 Mar 23;12:62. ISS results funded by a 2012 Space Biology NNH12ZTT001N grant nos. 19-12829-26 under Task Order NNN13D111T award to K.V., which also funded post-doctoral fellowship for N.K.S. K.R. acknowledges support from the Science and Engineering Board (SERB) MATRICS Grant MTR/2020/000490, IIT Madras, Centre for Integrative Biology and Systems mEdicine (IBSE) and Robert Bosch Center for Data Science and Artificial Intelligence (RBCDSAI).
Genomic, functional, and metabolic enhancements in multidrug-resistant Enterobacter bugandensis facilitating its persistence and succession in the International Space Station, Microbiome (open access)
Astrobiology