Enceladus Trails A Wake Of Electromagnetic Ripples That Extend Over Half A Million Kilometers
A major study by an international team of researchers using data from the NASA/ESA/ASI Cassini spacecraft has revealed a lattice-like structure of crisscrossing reflected waves that flow downstream behind the moon in Saturn’s equatorial plane, but also reach up to very high northern and southern latitudes.
The analysis of data from four instruments aboard Cassini, collected over the mission’s 13-year duration, demonstrates the crucial role that Enceladus plays in circulating energy and momentum around Saturn’s space environment.

Plumes of water vapour and dust stream through cracks in the icy surface of the southern hemisphere of Enceladus. The water molecules and particles from these geysers become ionised when exposed to radiation, creating an electrically-charged plasma that interacts with Saturn’s magnetic field as it sweeps past Enceladus. Credit: NASA/JPL/Space Science Institute.
“Enceladus, Saturn’s small icy moon, is famous for its water geysers, but its actual impact and interaction with the giant planet has remained partly unknown. This result from Cassini transforms our vision of the moon’s role in the Saturnian system,” said Lina Hadid of the Laboratoire de Physique de Plasmas (LPP) in France, who led the study.
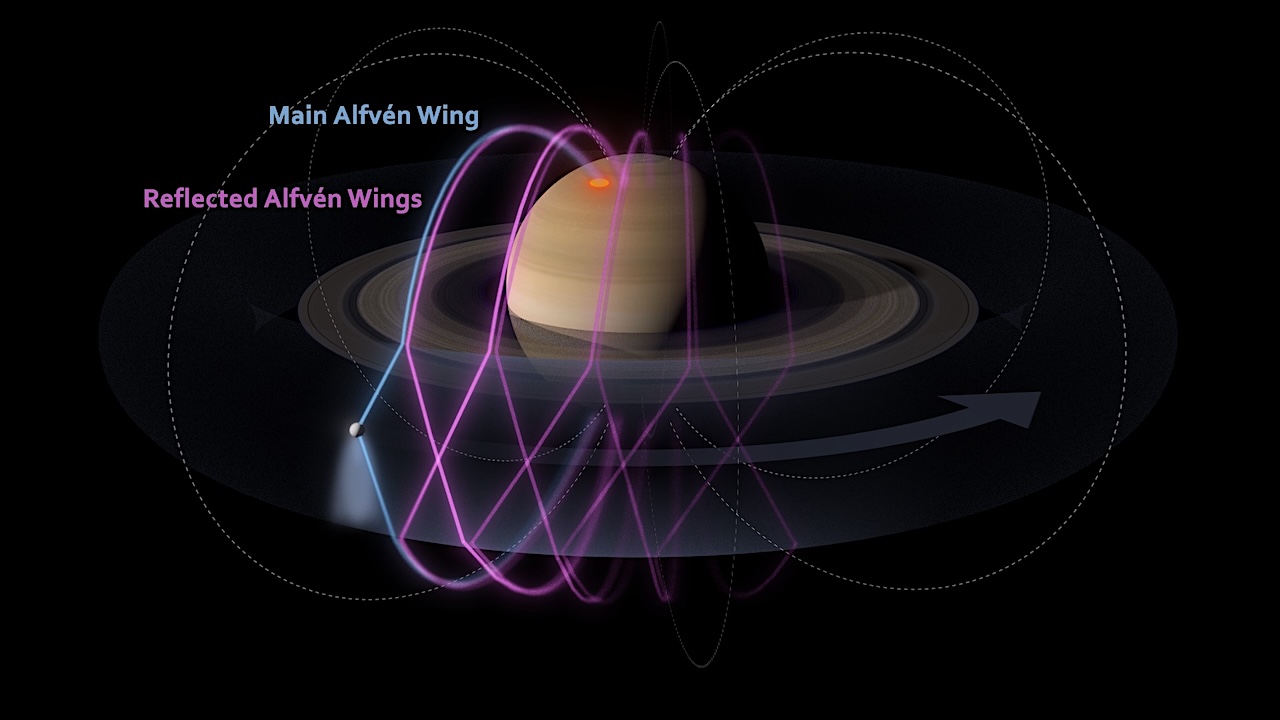
The study, published in the Journal of Geophysical Research: Space Physics, shows how wave structures, known as ‘Alfvén wings’, travel like vibrations on a string along magnetic field lines connecting Enceladus to Saturn’s pole. The initial ‘main’ Alfvén wing is reflected back-and-forth both by Saturn’s ionosphere and the plasma torus that encircles Enceladus’s orbit, resulting in a complex and structured system. By using a multi-instrumental approach, researchers were able to show that the influence of Enceladus extends over a record distance of over 504,000 km – more than 2,000 times the moon’s radius.
“This is the first time such an extensive electromagnetic reach by Enceladus has been observed, proving that this small moon acts as a giant planetary-scale Alfvén wave generator,” said Thomas Chust of LPP, co-author of the study. “This work sets the stage for future studies of other systems, such as the icy moons of Jupiter or exoplanets, by showing that a small moon with an electrically-conducting atmosphere can influence its host over vast distances on the scale of the giant planet itself.”
The researchers examined archive data from the suite of instruments carried by Cassini to study electromagnetic wave and particle interactions, looking for flyby and non-flyby paths near Enceladus that showed evidence of magnetic connections between the moon and Saturn. On 36 occasions, they found signatures related to Alfvén waves, including at much further distances than they originally anticipated.
As well as the large-scale structures, the team found evidence that turbulence teases out the waves into filaments within the main Alfvén wing. This fine-scale structure helps the waves bounce off Enceladus’s plasma torus and reach the high-latitudes in Saturn’s ionosphere where auroral features associated with the moon form.
“These results highlight the importance for future missions to Enceladus, such as the planned ESA orbiter and lander in the 2040s, to carry instrumentation that can study these electromagnetic interactions in even more detail,” said Hadid.
The study was led by LPP in collaboration with researchers from French laboratories including IRAP, ISAE-SUPAERO, LATMOS, LAM, and LIRA/Observatoire de Paris. International institutions participating in the study included ESA, IRFU in Sweden, MPS in Germany, CAS in the Czech Republic, Johns Hopkins APL, UCLA, the Universities of Michigan, Boston, and Iowa in the United States, DIAS in Ireland, MSSL/UCL, and Imperial College London in the United Kingdom. The CDPP/AMDA tool used in the study was supported through the Europlanet 2024 Research Infrastructure project with funding from the European Commission.
Publication
Hadid, L. Z., Chust, T., Wahlund, J.‐E., Morooka, M. W., Roussos, E., Witasse, O., et al. (2026). Evidence of an extended Alfvén wing system at Enceladus: Cassini’s multi‐instrument observations. Journal of Geophysical Research: Space Physics, 131, e2025JA034657. https://doi.org/10.1029/2025JA034657
Astrobiology,