Researchers Challenge A Fundamental Assumption In Evolutionary Biochemistry
How did life originate? Ancient proteins may hold important clues. Every organism on Earth is made up of proteins. Although all organisms — even single-celled ones — have complex protein structures now, this wasn’t always the case.
For years, evolutionary biochemists assumed that the most ancient proteins emerged from a simple signature, called a motif. New research from, though, suggests that this motif, without the surrounding protein, isn’t as consequential as it seemed. The international team of researchers was led by Lynn Kamerlin, a professor in the Georgia Tech School of Chemistry and Biochemistry and Georgia Research Alliance Vasser Woolley Chair in Molecular Design, and Liam Longo, a Specially Appointed Associate Professor at Earth-Life Science Institute (ELSI) at Institute of Science Tokyo, in Japan.
“It’s probably an eroded molecular fossil, with its true nature having been overwritten over billions of years of evolution,” said Kamerlin. “This work completely reshapes how we think about proteins. It’s like trying to play protein Jeopardy! — now we need to rethink what the original question was.”
Prehistoric Proteins
It’s not hard to understand why this hypothesis was wrong for so long. The motif is associated with the element phosphorus, one of the key elements of life. Many of the earliest proteins bound to phosphorus-containing compounds. While these early proteins have different structures, they frequently share the same motif.
“For years, researchers took this to mean that today’s complex proteins came from the motif itself — that this tiny protein gave rise to entire families,” Longo said.
To discover the protein’s origins, the researchers pored over available data on protein crystal structures. Then they identified and characterised relevant proteins computationally. Although they recognised some of the protein’s similar structure in their modelling, the motif was not identical. They found that many different types of phosphate-binding proteins were possible. The idea that this motif was somehow special on its own was false.
“We don’t hypothesise that eyes gave rise to heads, even though nearly all heads have eyes; that’s because seeing involves interlocking systems,” Kamerlin said. “Our early peptide presents a similar instance. Only embedding within the larger system allows it to shine.”
Protein Possibilities
The researchers tested this work in water and methanol environments. Methanol mimics environments on Earth that may have less water around. The researchers found comparable protein motifs in this methanol environment, proving that the famous motif was not unique, but rather one of many possible motifs with similar properties. What was assumed to be a building block of early life is probably just a fossil fragment — and not the complete picture.
Kamerlin and Longo’s work helps their field determine not just how life started but also bolsters biotechnology advancements. A better understanding of how natural proteins evolved will help other researchers create artificial proteins, for everything from drug delivery to new vaccines.
The work is far from finished. Now that the researchers know this protein motif is one of many possible options, the question becomes: When did this motif become dominant, and what else could life have looked like? These questions will help the scientific world make discoveries that could benefit everyone.
Funding from the Knut and Alice Wallenberg Foundation; the Okinawa Institute of Science and Technology Graduate University (OIST) with subsidy funding from the Cabinet Office, Government of Japan; and the National Academic Infrastructure for Supercomputing in Sweden.
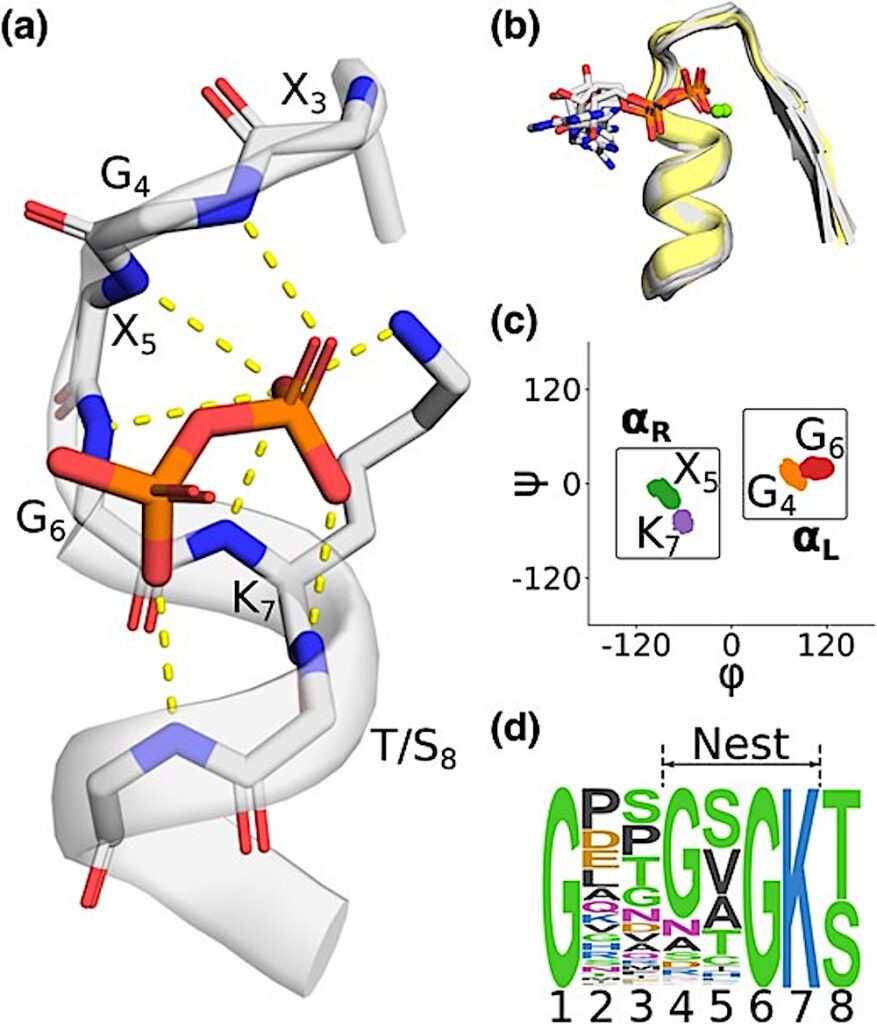
The phosphate binding nest of P-Loop NTPases. a) A representative phosphate binding nest, with 6 backbone amides and one lysine side chain oriented toward and interacting (dashed lines) with the pyrophosphate moiety of ADP. ECOD domain e3k1jA2. b) Structural context of the Walker A motifs analyzed in this study. Structures with a bound phospho-ligand are colored gray; unbound structures are colored yellow. Mg2+ ions are drawn as green spheres and are present in only a subset of structures. Mg2+ ions were not included in our simulations, as described in the main text. ECOD domains: e3ievA2, e2it1A8, e3k1jA2, e5jrjA2, e6ojxA1, e2qy9A2. Note that the γ phosphate of the bound ATP was removed for clarity in e6ojxA1. c) Ramachandran plot showing the per residue distribution of ϕ, ψ angles, including both liganded and unliganded structures. Boxes indicate the regions taken to be ɑR and ɑL in subsequent analyses. Note the alternating ɑL-ɑR-ɑL-ɑR character of the nest. d) Sequence logo of the Walker A Motif demonstrating the canonical GxxGxGK[T/S] sequence pattern. P-loop NTPase sequences were gathered from the PDB and filtered for 90% identity at the domain level. — Molecular Biology and Evolution
Redefining the Limits of Functional Continuity in the Early Evolution of P-Loop NTPases, Molecular Biology and Evolution (open access)
Astrobiology,