Recoding A Genome For Programmable Synthetic Proteins
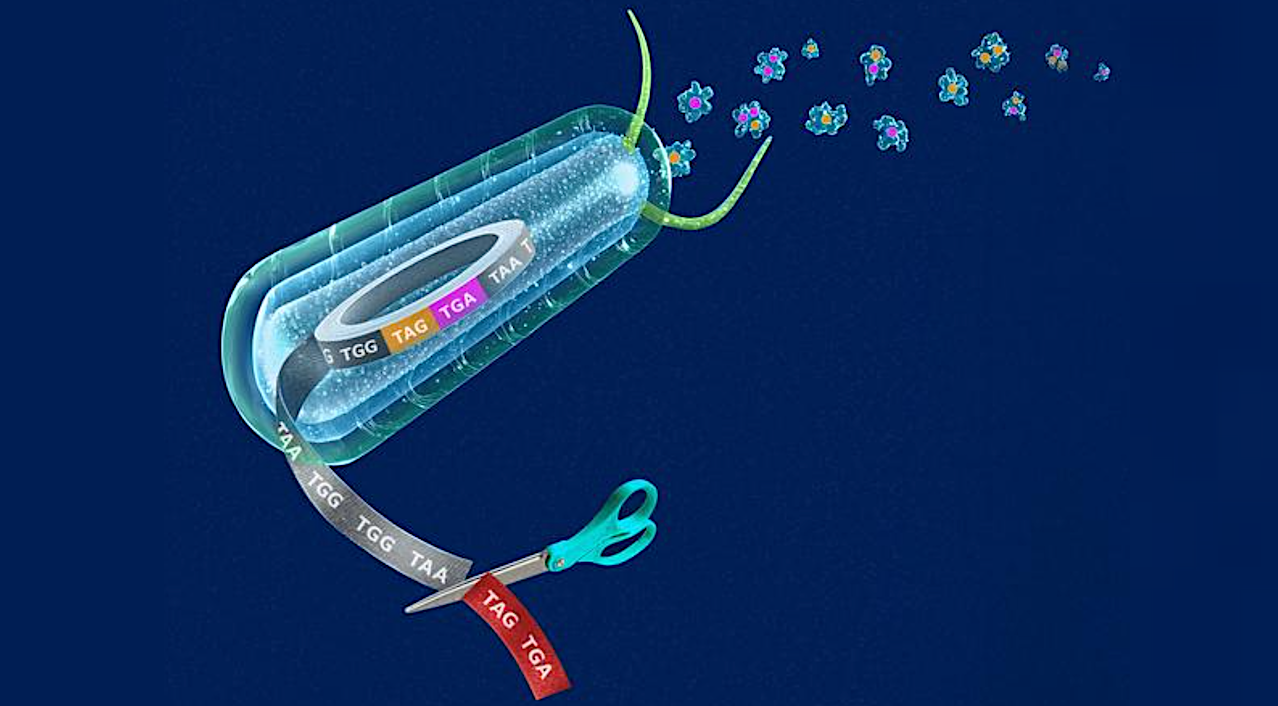
Synthetic biologists from Yale were able to re-write the genetic code of an organism — a novel genomically recoded organism (GRO) with one stop codon — using a cellular platform that they developed enabling the production of new classes of synthetic proteins. These synthetic proteins, researchers say, offer the promise of innumerable medical and industrial applications that can benefit society and human health.
The creation of the landmark GRO, known as “Ochre” — which fully compresses redundant, or “degenerate” codons, into a single codon — is described in a new study published Feb. 5 in the journal Nature. A codon is a sequence of three nucleotides in DNA or RNA that codes for a specific amino acid, which serve as the biochemical building blocks for proteins.
“This research allows us to ask fundamental questions about the malleability of genetic codes,” said Farren Isaacs, professor of molecular, cellular and developmental biology at Yale School of Medicine and of biomedical engineering at Yale’s Faculty of Arts and Sciences, who is co-senior author of the paper. “It also demonstrates the ability to engineer the genetic code to endow multi-functionality into proteins and usher in a new era of programmable biotherapeutics and biomaterials.”
The landmark advance builds on a 2013 study by the team, published in Science, which described the construction of the first GRO. In that study, the researchers demonstrated new solutions for safeguarding genetically engineered organisms and for producing new classes of synthetic proteins and biomaterials with “unnatural,” or human-created, chemistries.
Ochre is a major step toward creating a non-redundant genetic code in E. coli, specifically, which is ideally suited to produce synthetic proteins containing multiple, different synthetic amino acids.
Jesse Rinehart, an associate professor of cellular and molecular physiology at the Yale School of Medicine and co-senior author on the study, called the breakthrough a “profound piece of whole genome engineering based on over 1,000 precise edits at a scale an order of magnitude greater than any engineering feat we have previously done.”
“This is an exciting new platform technology that opens up an array of applications for biotechnology both in the academic realm and in the commercial sector,” Rinehart said. “We want to advance our general knowledge of science but we also want to enable industrial applications that are beneficial to society.”
The codon, a sequence of three nucleotides in DNA or RNA, acts like an “instruction manual” for protein synthesis, telling the cell which of the 20 natural amino acids to add to a growing protein chain (or, in the case of “stop” codons, signaling the termination of protein synthesis). In this process, known as translation, the genetic information carried in a messenger RNA (mRNA), via the genetic code, dictates not only the order of amino acids but also when the process should start and stop.
Michael Grome, a postdoctoral associate in molecular, cellular, and developmental biology at Yale and first author of the study, likened codons to three-letter words within a sentence in the genetic recipe for life. Inside the cell, he said, there are ribosomes that act like 3-D printers that read the recipe. Each word calls for one “ingredient” amino acid from among the list of 20 natural amino acids that make up proteins.
“A lot of these words are equivalent, or synonymous,” Grome said. “We set out to add more ingredients for building proteins, so we took three of these words for ‘stop’ and made them one. Two words were removed, then we re-engineered the cell so they were ‘freed’ for new function. We then engineered a cell that recognized the word to say something new, to represent a new ingredient.”
Specifically, the researchers eliminated two of the three stop codons that terminate protein production. The recoded genome reassigned four codons to non-degenerate functions, including the two recoded stop codons dedicated to encoding nonstandard, or unnatural, amino acids into protein.
In addition to introducing thousands of precise edits across the genome, the work required AI-guided design and re-engineering of essential protein and RNA translation factors to create a strain capable of adding two nonstandard amino acids into its recipe book. These nonstandard amino acids imbue proteins with multiple new properties, such as programmable biologics with reduced immunogenicity (a substance’s ability to induce an immune response in the body) or biomaterials with enhanced conductivity.
The results reflect years of recoding work by the two labs at the Yale Systems Biology Institute on West Campus. The collaboration between Rinehart and Isaacs dates to 2010 when they began working in neighboring labs. Isaacs has long been interested in engineering genomes — much like, he said, an architect might plan and make changes to a building. Rinehart’s work focuses on proteins — how they are made and how the stage might be set for them to carry out other actions.
“We recognized we have complementary expertise and that both labs bring a broad set of expertise and capability,” Rinehart said.
Isaacs is excited about what he describes as the potentially “killer” applications for programmable protein biologics that the new platform will make possible. One such application involves engineering protein drugs with synthetic chemistries to decrease the frequency of dosing or undesirable immune responses. The team reported such an application using their first-generation GRO in a 2022 study. In that study they encoded non-standard amino acids into protein, demonstrating a safer, controllable approach to precisely tune the half-life of protein biologics.
The new Ochre cell expands these capabilities for use in the construction of multi-functional biologics. Isaacs and Rinehart are currently acting as advisors to Pear Bio, a Yale biotechnology spin-off that has licensed the technology for commercializing programmable biologics.
Other Yale researchers involved in the study are Michael Nguyen, Daniel Moonan, Kyle Mohler, Kebron Gurara, Shenqi Wang, Colin Hemez, Benjamin Stenton, Yunteng Cao, Felix Radford, Maya Kornaj, Jaymin Patel, Maisha Prome, Svetlana Rogulina, David Sozanski, and Jesse Tordoff.
Engineering a genomically recoded organism with one stop codon, Nature (open access)
Astrobiology, Genomics,