Ancient Microbes Offer Clues To How Complex Life Evolved
A new study published in Science Advances reveals a surprising twist in the evolutionary history of complex life. Researchers at Queen Mary University of London have discovered that a single-celled organism, a close relative of animals, harbors the remnants of ancient giant viruses woven into its own genetic code. This finding sheds light on how complex organisms may have acquired some of their genes and highlights the dynamic interplay between viruses and their hosts.
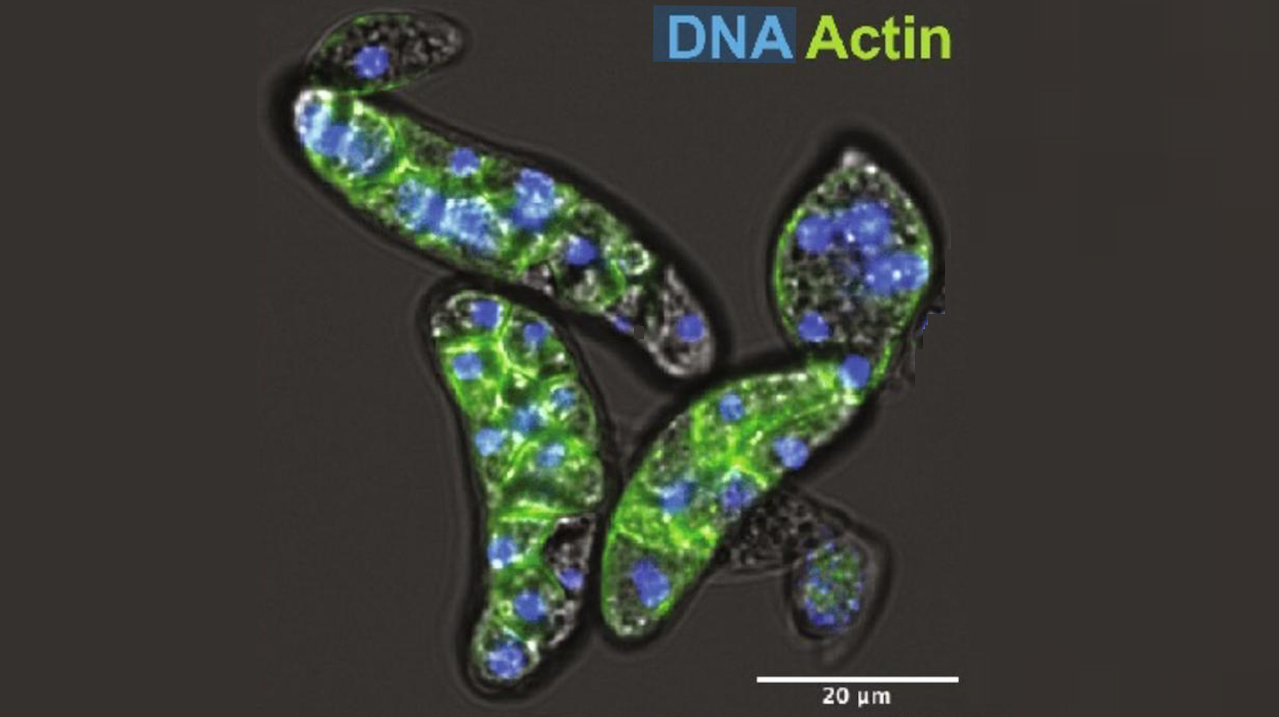
The study focused on a microbe called Amoebidium, a unicellular parasite found in freshwater environments. By analysing Amoebidium’s genome, the researchers led by Dr Alex de Mendoza Soler, Senior Lecturer at Queen Mary’s School of Biological and Behavioural Sciences, found a surprising abundance of genetic material originating from giant viruses – some of the largest viruses known to science. These viral sequences were heavily methylated, a chemical tag that often silences genes.
“It’s like finding Trojan horses hiding inside the Amoebidium’s DNA,” explains Dr de Mendoza Soler. “These viral insertions are potentially harmful, but Amoebidium seems to be keeping them in check by chemically silencing them.”
The researchers then investigated how widespread this phenomenon might be. They compared the genomes of several Amoebidium isolates and found significant variation in the viral content. This suggests that the process of viral integration and silencing is ongoing and dynamic.
“These findings challenge our understanding of the relationship between viruses and their hosts,” says Dr. de Mendoza Soler. “Traditionally, viruses are seen as invaders, but this study suggests a more complex story. Viral insertions may have played a role in the evolution of complex organisms by providing them with new genes. And this is allowed by the chemical taming of these intruders DNA.”
Furthermore, the findings in Amoebidium offer intriguing parallels to how our own genomes interact with viruses. Similar to Amoebidium, humans and other mammals have remnants of ancient viruses, called Endogenous Retroviruses, integrated into their DNA. While these remnants were previously thought to be inactive “junk DNA,” some might now be beneficial. However, unlike the giant viruses found in Amoebidium, Endogenous Retroviruses are much smaller, and the human genome is significantly larger. Future research can explore these similarities and differences to understand the complex interplay between viruses and complex life forms.

A. appalachense displays a methylome with gene body and transposon methylation. (A) Distribution of DNMTs in eukaryotes and teretosporeans. Black dot indicates enzyme presence, white dot indicates enzyme absence, and black triangles indicate species for which Enzymatic Methyl-seq has been performed. Amoebidium cells are stained with phalloidin and Hoechst. Phylogenetic relationships based on previous studies (29). (B) Distribution of genome sizes, repetitive content, and global methylation levels across opisthokonts. Asterisks indicate species for which the lack of 5mC is inferred from their absence of DNMTs. Genome sizes for species with genomes above 400 Mb are displayed in white, and repeat content (%) is also highlighted for clarity. (C) Gene body methylation and (D) TE methylation averages split by the extended CG sequence context. Not expressed genes display <1 TPM. TSS, transcriptional start site; TES, transcriptional end site. TEs only include copies spanning at least 70% of the RepeatModeler2 consensus model. — Queen Mary University of London
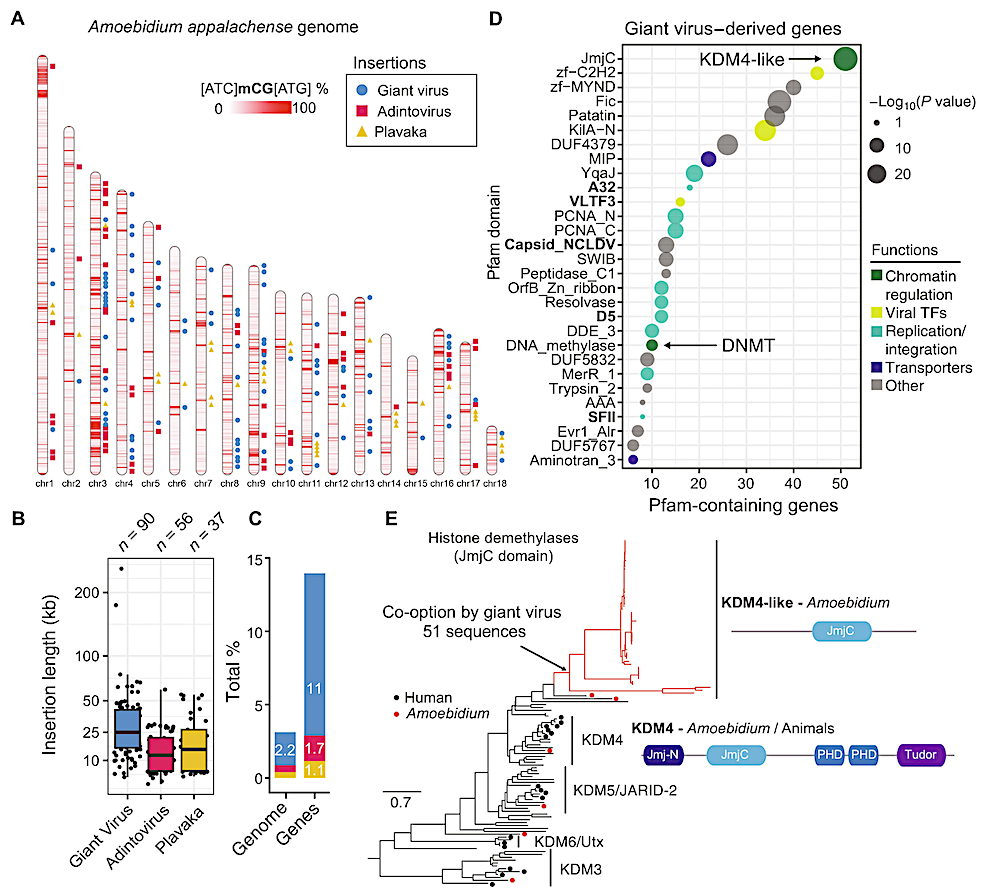
The Amoebidium genome harbors hundreds of viral insertions. (A) Location of GEVEs, adintovirus, and Plavaka giant repeats across the Amoebidium genome. Windows of 10 kb are colored according to their methylation level in non-CGC/GCG trinucleotides. (B) Distribution of insertion sizes of the giant repeats within chromosomes. Center lines in boxplots are the median, box is the interquartile range (IQR), and whiskers are the first or third quartile ± 1.5× IQR. (C) Contribution of giant repeats to genome size and gene counts. (D) Pfam domains enriched in genes encoded in GEVE regions. In bold, marker giant virus domains. The displayed P values correspond to a two-sided Fisher exact test. (E) Maximum likelihood phylogeny of JmjC in eukaryotes, highlighting the expansion of KDM4-like enzymes in GEVE regions. Black dots indicate human sequences, red dots indicate Amoebidium sequences, and red branches indicate genes within endogenized viral regions. Domain architectures defined with PFAM domains. — Queen Mary University of London
DNA methylation enables recurrent endogenisation of giant viruses in an animal relative, Science Advances (open access)
Astrobiology,