Development of Allosteric Ribozymes for ATP and l-Histidine Based on the R3C Ligase Ribozyme
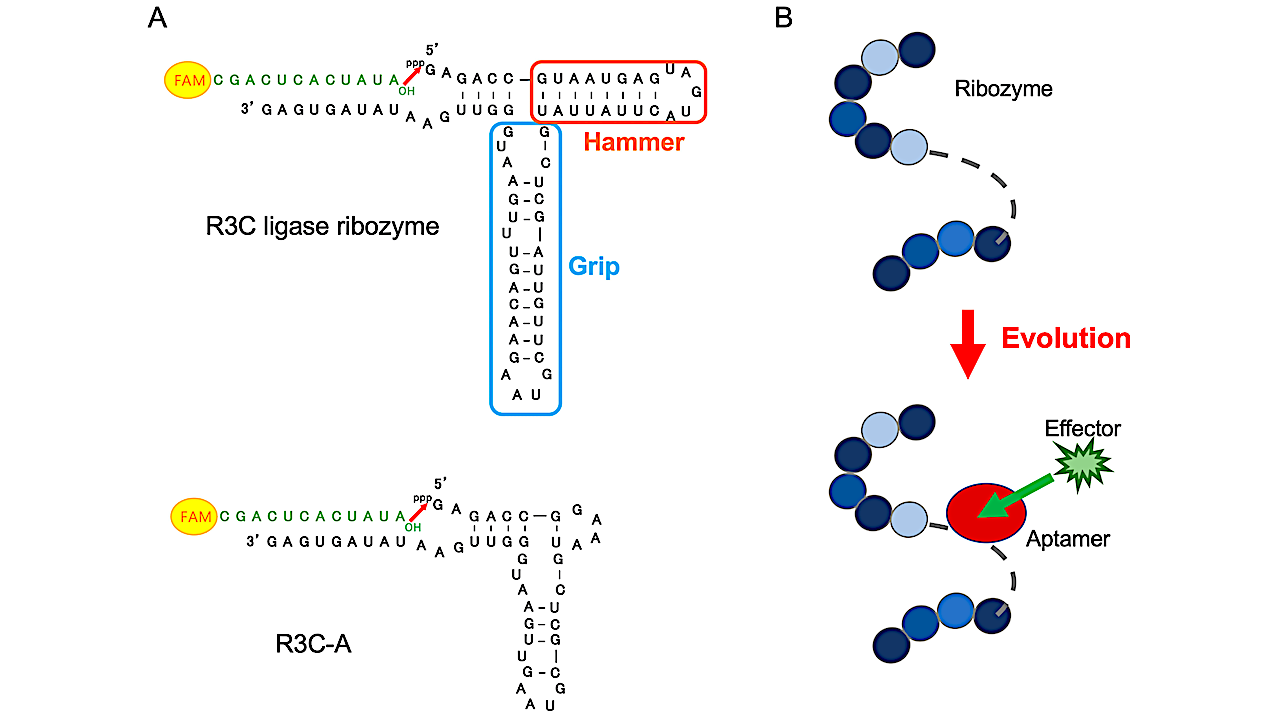
During the evolution of the RNA, short RNAs are thought to have joined together to form long RNAs, enhancing their function as ribozymes.
Previously, the artificial R3C ligase ribozyme (73 nucleotides) was successfully reduced to 46 nucleotides; however, its activity decreased significantly. Therefore, we aimed to develop allosteric ribozymes, whose activities could be regulated by effector compounds, based on the reduced R3C ligase ribozyme (R3C-A). Among the variants prepared by fusing an ATP-binding aptamer RNA with R3C-A, one mutant showed increased ligation activity in an ATP-dependent manner.
Melting temperature measurements of the two RNA mutants suggested that the region around the aptamer site was stabilized by the addition of ATP. This resulted in a suitable conformation for the reaction at the ligation site. Another ribozyme was prepared by fusing R3C-A with a l-histidine-binding aptamer RNA, and the ligase activity increased with increasing l-histidine concentrations. Both ATP and l-histidine play prominent roles in current molecular biology and the interaction of RNAs and these molecules could be a key step in the evolution of the world of RNAs.
Our results suggest promise in the development of general allosteric ribozymes that are independent of the type of effector molecule and provide important clues to the evolution of the RNA world.
Yuna Akatsu, Hiromi Mutsuro-Aoki and Koji Tamura
Life 2024, 14(4), 520; DOI: 10.3390/life14040520
Development of Allosteric Ribozymes for ATP and l-Histidine Based on the R3C Ligase Ribozyme, Life (open access)
Astrobiology