Transcriptome Analysis Of The Tardigrade Hypsibius Exemplaris Exposed To The DNA-damaging Agent Bleomycin
Tardigrades are microscopic animals that are renowned for their capabilities of tolerating near-complete desiccation by entering an ametabolic state called anhydrobiosis.
However, many species also show high tolerance against radiation in the active state as well, suggesting cross-tolerance via the anhydrobiosis mechanism. Previous studies utilized indirect DNA damaging agents to identify core components of the cross-tolerance machinery; however, it was difficult to distinguish whether transcriptomic changes were the result of DNA damage or residual oxidative stress.
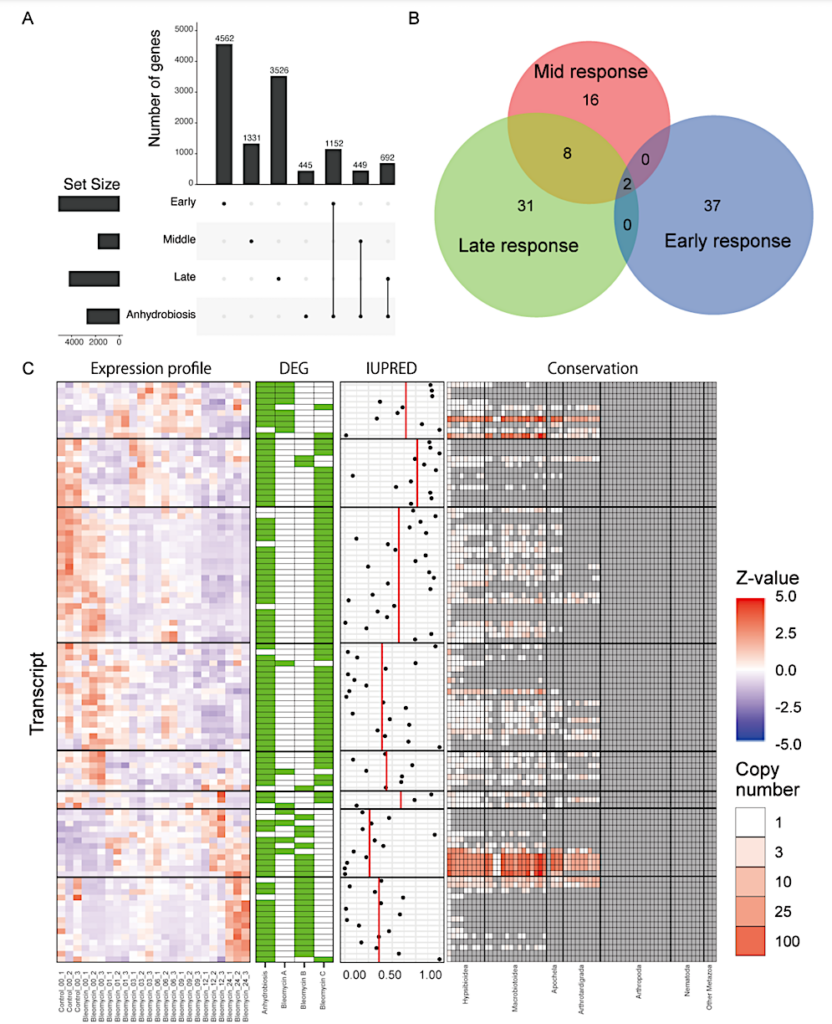
Comparison with anhydrobiosis entry. [A] UpSet plot showing the number of genes shared between each cluster and anhydrobiosis entry. [B] Number of GO BP terms significantly enriched between each response cluster and anhydrobiosis. [C] Features of the 102 NLS predicted transcripts without annotations, induced in anhydrobiosis and either one of lusters A, B, C. Transcripts are ordered based on the clustering preformed Figure S1C. Z-scaled expression profiles, DEG profiles, IUPRED3 score, conservation patterns across Tardigrada and relative lineages are shown. — biorxiv.org
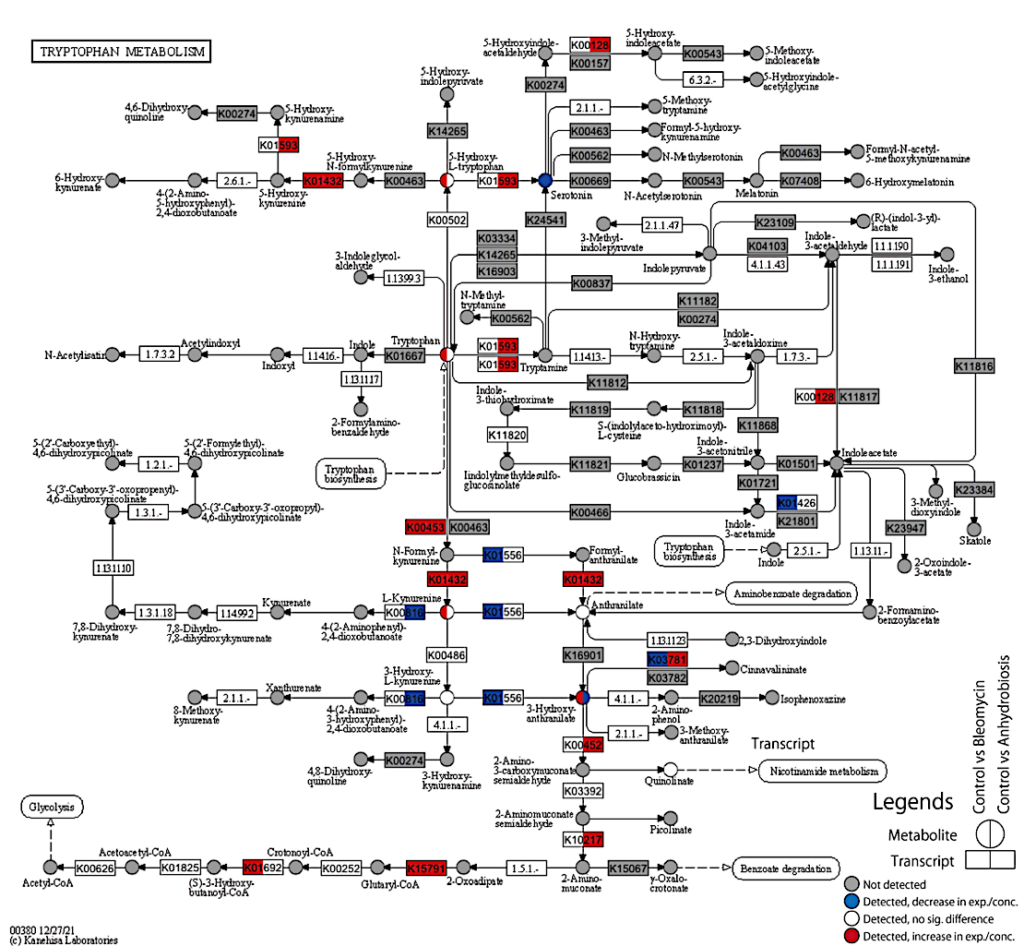
KEGG pathway map of tryptophan metabolism. Data for two comparisons are indicated in a single circle (metabolite) or square (transcript); For metabolites, Left: Control vs Bleomycin; Right : Control vs Anhydrobiosis; For transcripts; Left: Control vs Bleomycin-00h (24h exposure, no recovery time), Right: Active vs Tun. Colors correspond to the following conditions: Gray: not detectable or not detected (0µg/µL for each condition), Blue: decrease in expression/concentration (lower in Bleomycin/Anhydrobiosis); White: detected, but no significant difference; Red: increase in expression/concentration (higher in Bleomycin/Anhydrobiosis).– biorxiv.org
To this end, we performed transcriptome analysis on bleomycin-exposed Hypsibius exemplaris. We observed induction of several tardigrade-specific gene families that may be the core components of the cross-tolerance mechanism. We also identified an enrichment of the tryptophan metabolism pathway, which metabolomic analysis suggested the engagement of this pathway in stress tolerance. These results provide several candidates for the core component of the cross-tolerance, as well as possible anhydrobiosis machinery.
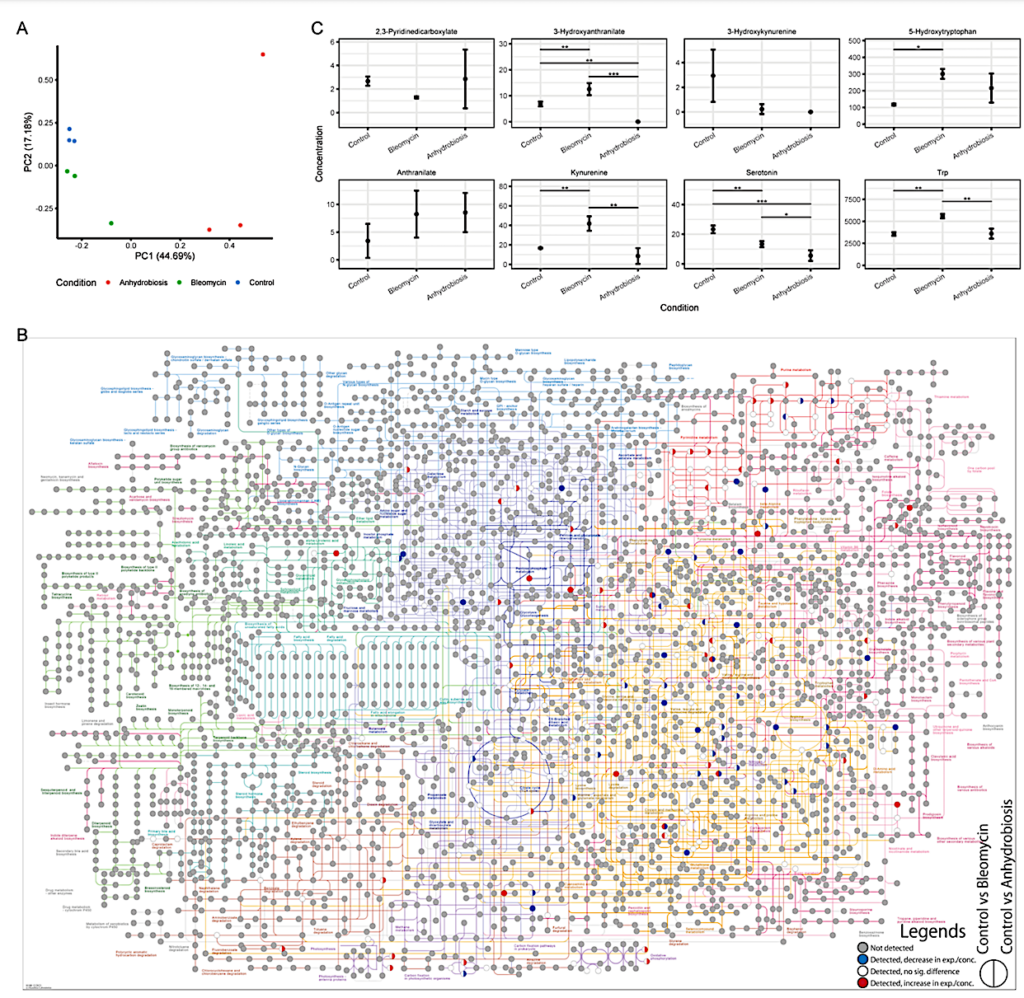
General analysis on metabolome data. [A] PCA of metabolome compound concentrations. [B] KEGG pathway colored with metabolite profiles. Data for two comparisons are shown in a single circle Left: Control vs Bleomycin; Right : Control vs Anhydrobiosis. Colors correspond to the following conditions: Colors correspond to the following conditions: Gray: not detectable or not detected (0µg/µL for each condition), Blue: decrease in expression/concentration (lower in Bleomycin/Anhydrobiosis); White: detected, but no significant difference; Red: increase in expression/concentration (higher in Bleomycin/Anhydrobiosis) [C] Concentration profiles for trytophan metabolites. — biorxiv.org
Yuki Yoshida, Akiyoshi Hirayama, Kazuharu Arakawa
Transcriptome analysis of the tardigrade Hypsibius exemplaris exposed to the DNA-damaging agent bleomycin, biorxiv.org
doi: https://doi.org/10.1101/2024.02.01.578372
Astrobiology