Microbial Wonders In Earth’s Saltiest Waters
The study delves into hypersaline lakes in Xinjiang, China, exploring the genetic and metabolic diversity of microbial communities termed “microbial dark matters”.
Hypersaline lake ecosystems, characterized by extreme salinity, harbor unique microorganisms with largely unexplored biosynthesis and biodegradation capabilities. The research seeks to uncover novel biological compounds and pathways, potentially revolutionizing biotechnology, medicine, and environmental remediation by tapping into the untapped potential of these extremophiles.
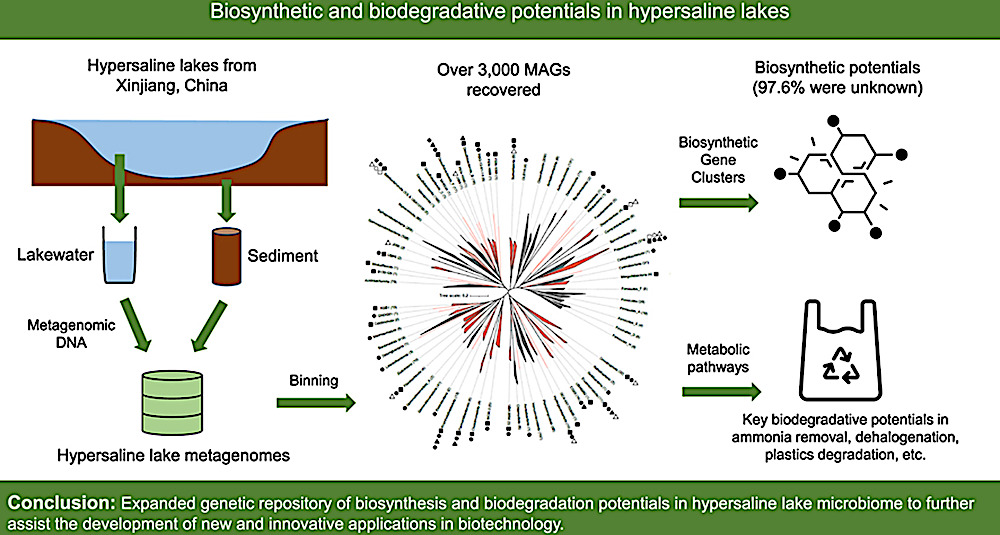
Graphical abstract. CREDIT Environmental Science and Ecotechnology / Science Direct
A recent study (https://doi.org/10.1016/j.ese.2023.100359) published in Volume 20 of the journal Environmental Science and Ecotechnology, explores the largely unknown metabolic capabilities of unclassified microbial species in extreme environments, particularly hypersaline lakes, and their potential applications in biotechnology, medicine, and environmental remediation.
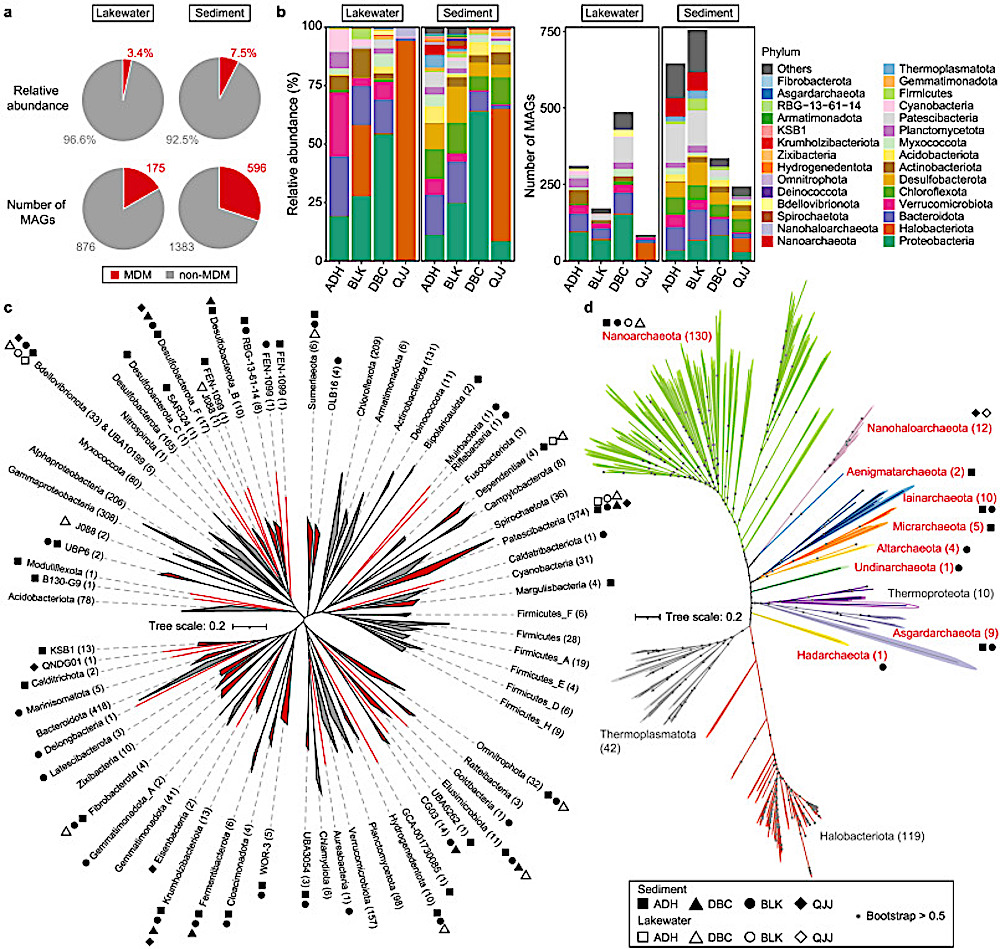
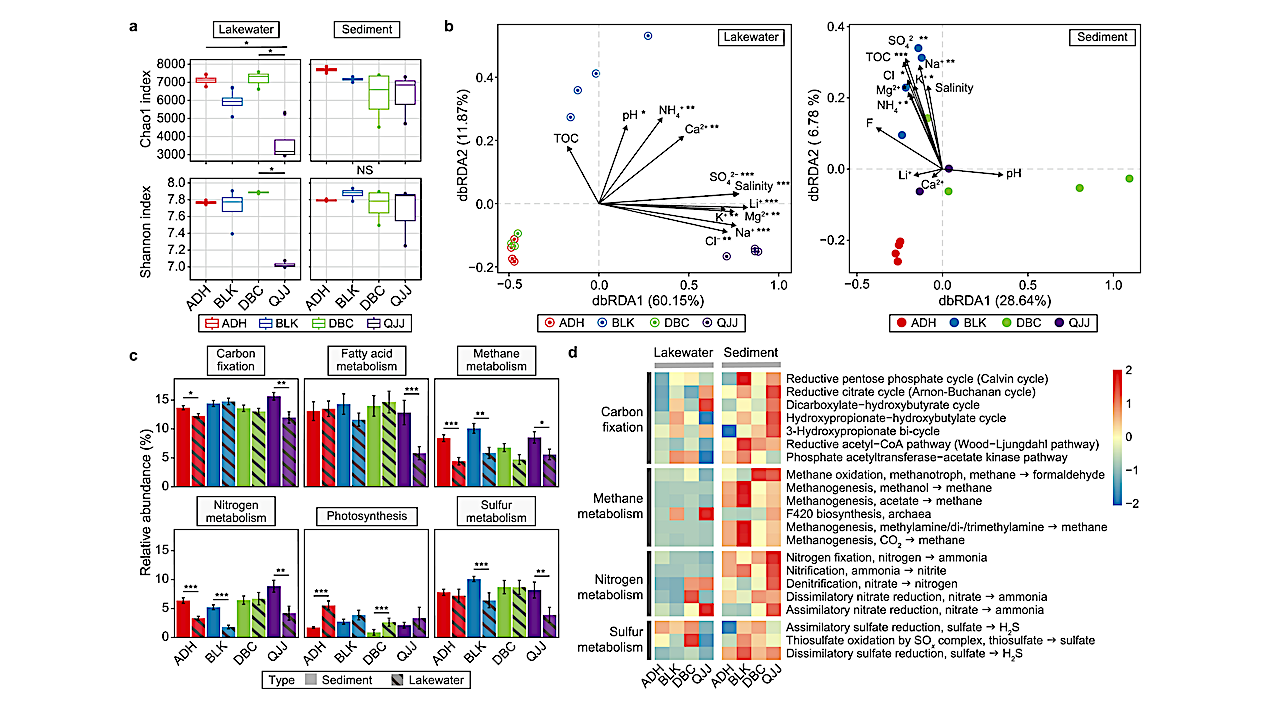
Phylogenetic analysis of MDM MAGs. Unrooted maximum likelihood trees of archaeal and bacterial MAGs were constructed with 122 and 120 concatenated marker genes in IQ-TREE using Q.pfam+R10 and LG+F+R10 models, respectively. a, MDM proportion of lakewater and sedimentary communities in terms of relative abundance and number of MAGs. Red: MDMs; Grey: non-MDMs. b, Microbial structure of lakewater and sedimentary communities in different lakes regarding relative abundance and number of MAGs classified at the phylum level. c–d, Phylogenetic tree of bacterial (c) and archaeal (d) MAGs. MDM lineages (bacterial) or phyla (archaeal) were highlighted in red, and MDM MAG(s) present in the corresponding lineages/phyla were indicated with shapes. ADH: square, BLK: circle, DBC: triangle, QJJ: diamond, solid symbol: sediment, hallowed symbol: plankton. CREDIT Environmental Science and Ecotechnology / Science Direct
In this detailed study, researchers embarked on a scientific adventure to the salt-rich lakes of Xinjiang, China, aiming to explore the largely unknown world of microbial dark matters. These are microbes that thrive in environments with high levels of salt, which have not been classified due to their elusive nature. Utilizing advanced DNA sequencing techniques, the team cataloged an astonishing variety of over 3,000 metagenome-assembled genomes (MAGs) from 82 different families, most of which are new to science. They unearthed more than 9,000 unique biosynthesis gene clusters, 94% of which are novel, indicating a vast, untapped potential for new biological discoveries. This research not only expands our knowledge of life in extreme conditions but also opens exciting possibilities for new technologies and medical breakthroughs, leveraging the untapped resources of these unique microbial communities.
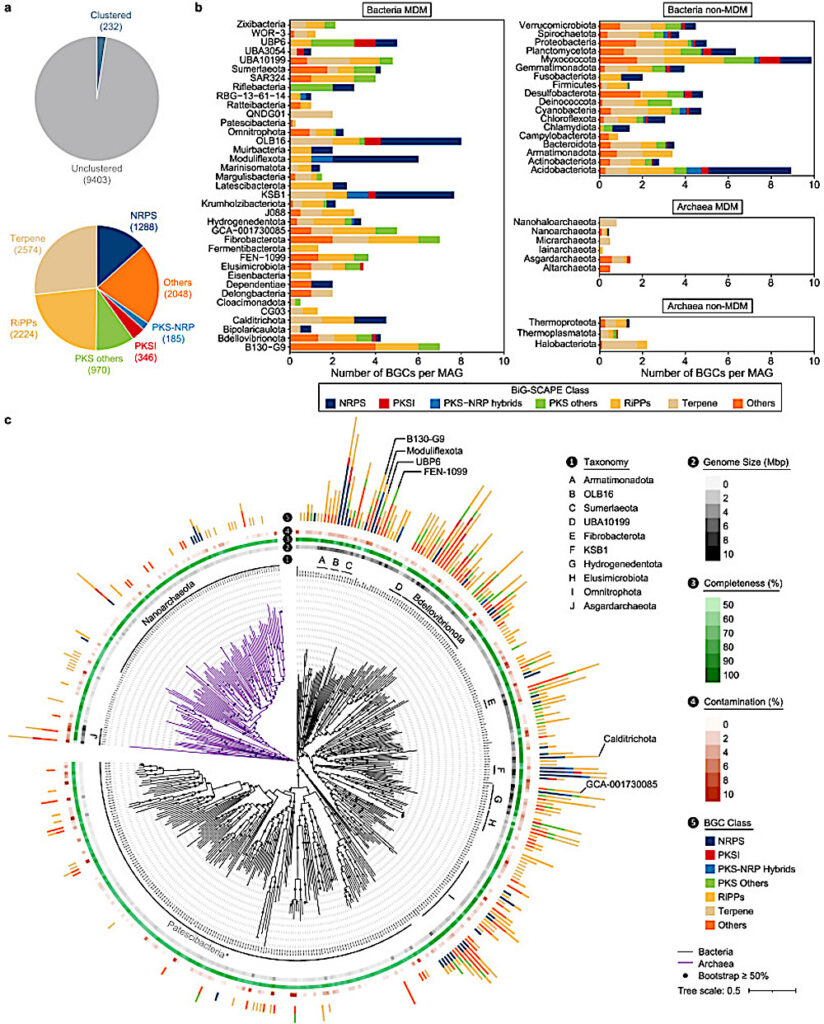
Analysis of biosynthetic potentials of hypersaline lake MAGs. a, A summary of predicted BGCs. In the pie chart on top, blue indicates that predicted BGCs were clustered to known BGCs in the MiBIG database; grey indicates that BGCs were not clustered to a known BGC in the MiBIG database. The pie chart at the bottom summarised the number of BGCs annotated to a BGC class. b, Normalised number of BGCs per MAG in each phylum. c, Phylogenetic trees of archaeal and bacterial MAGs with completeness ≥70, or number of BGCs ≥5. Midpoint-rooted maximum likelihood trees of archaeal and bacterial MAGs were constructed with 122 and 120 concatenated marker genes in IQ-TREE using Q.pfam+R10 and LG+F+R10 models, respectively. Black branches indicate bacterial MAGs, while purple branches indicate archaeal MAGs. CREDIT Environmental Science and Ecotechnology / Science Direct
Highlights
- Over 3000 MAGs were obtained from hypersaline lakes that enriched genomic resources.
- Microbial communities were significantly diversified across four hypersaline lakes.
- We identified 8000+ potential biosynthetic gene clusters in uncultured microbes.
- We uncovered biodegradation potential in several microbial dark matter lineages.
Ke Yu, the study’s lead researcher, emphasized the significance of these discoveries for biotechnology and environmental remediation, highlighting the untapped potential of microbial dark matters in extreme environments.
The findings open new avenues for biotechnological innovation, emphasizing the untapped potential of microbial diversity in extreme environments. The discovery of novel biosynthesis pathways and biodegradation capabilities in these microbial communities can have far-reaching implications for developing new drugs, biotechnological processes, and environmental remediation methods.
Astrobiology,