Getting Ready For Offworld Astrobiology Expeditions: A Universal DNA Barcode For The Tree Of Life
Editor’s note: According to this study “Species identification using DNA barcodes has revolutionized biodiversity sciences and society at large.“ When we start to explore inhabited worlds we’re going to have to tackle an entire world’s ecology. There may be a little biodiversity or a lot. And what we understand as “biodiversity” may, in and of itself, differ from what we associated with “life as we know it” on our world. Having the ability to organize classifications of the life forms we find would clearly be of great use. Developing data and systematics capabilities so as to be prepared for this herculean task that not only allow the classification of life forms – but also allow genetic relationships, evolution, and other genomic factors to be included – would be optimal. Here is one example of what has been proposed for all earthlings.
Species identification using DNA barcodes has revolutionized biodiversity sciences and society at large. However, conventional barcoding methods do not reflect genomic complexity, may lack sufficient variation, and rely on limited genomic loci that are not universal across the Tree of Life.
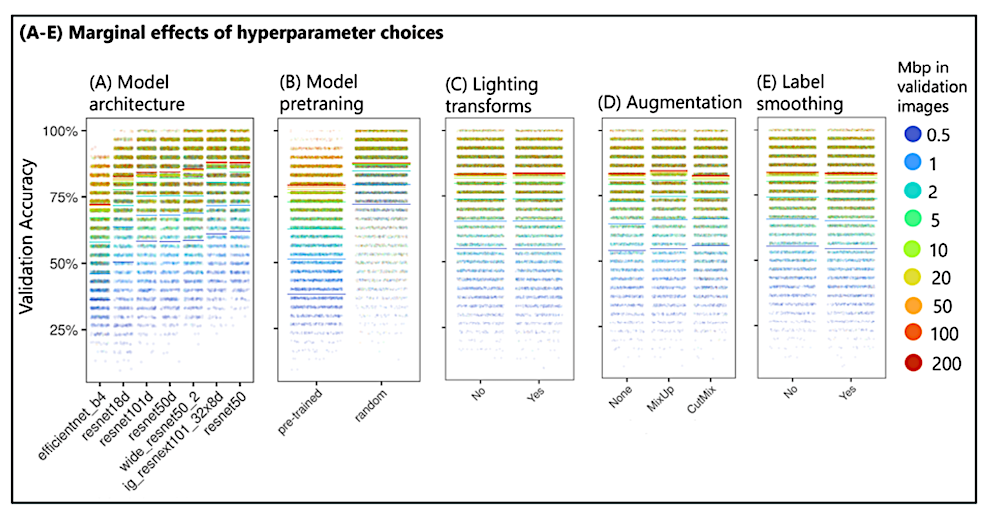
Marginal effects of neural network model and training options. Dots represent individual replicates, and bars depict averages. All parameters were identified to be significant in a linear model: more complex model architectures, lighting transformations, and augmentation methods MixUp and CutMix improved accuracy. However, pretraining with large image datasets and label smoothing decreased accuracy. — ecoevorxiv.org
Here, we develop a novel barcoding method that uses exceptionally low-coverage genome skim data to create a “varKode”, a two-dimensional image representing the genomic landscape of a species. Using these varKodes, we then train neural networks for precise taxonomic identification. Applying an expertly annotated genomic dataset including hundreds of newly sequenced genomic samples from the plant clade Malpighiales, we demonstrate >91% precision when identifying species or genera.
Remarkably, high accuracy remains despite minimal data amounts that lead to failure when applying alternative methods. We further illustrate the broad utility of varKodes across several focal clades of eukaryotes and prokaryotes. As a final test, we classify the entire NCBI eukaryote sequence-read archive to identify its 861 constituent families with >95% precision despite utilizing less than 10 Mbp of data per sample. Enhanced computational efficiency and scalability, minimal data inputs robust to degraded DNA, and modularity for further development make varKoding an ideal approach for biodiversity science.
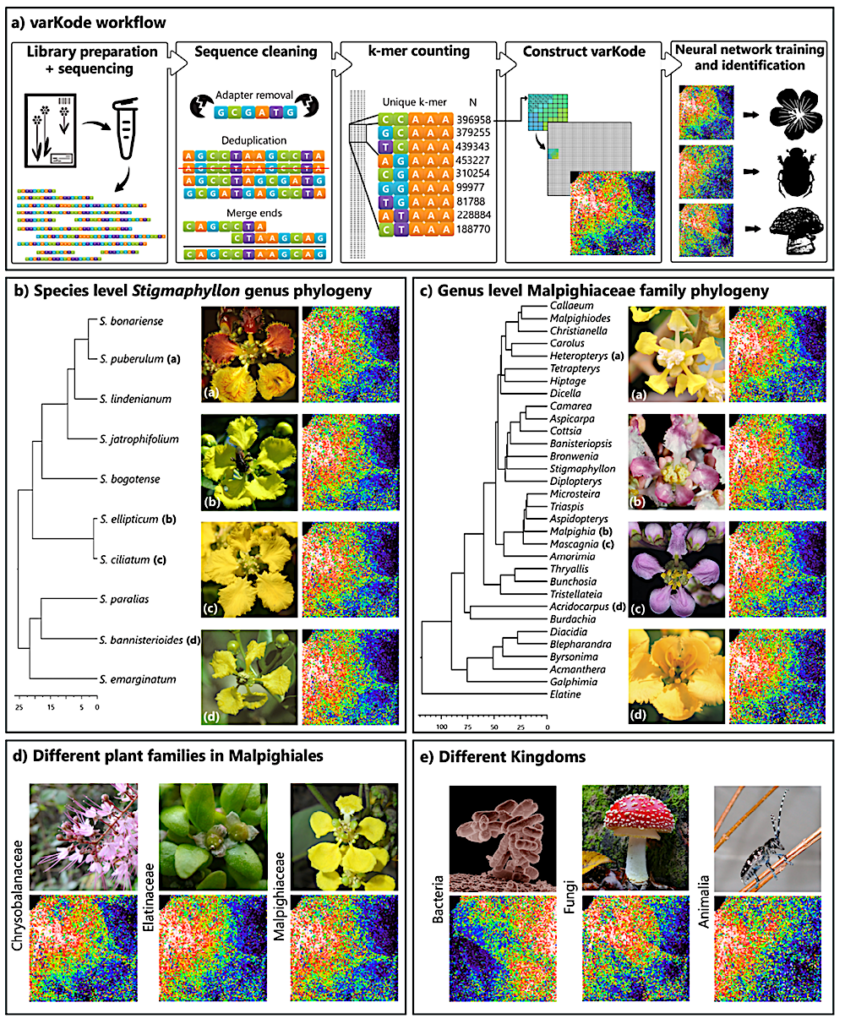
varKoding and training data overview. (A) varKode generation workflow. varKode images are natively grayscale, but here they are mapped to a rainbow color scale for increased contrast. (B) Phylogeny and example varKodes of Stigmaphyllon species. (C) Phylogeny and example varKodes of Malpighiaceae genera including their closest outgroup (Elatine, Elatinaceae). (D) Examples of varKodes from across plant families of Malpighiales, and (E) across kingdoms. Chronograms depicted for each representative set with timelines in millions of years (Myr) at the bottom of B and C. — ecoevorxiv.org
Bruno A S de Medeiros, Liming Cai, Peter J Flynn, Yujing Yan, Xiaoshan Duan, Lucas C Marinho, Christiane Anderson, Charles Davis
https://ecoevorxiv.org/repository/view/6567/
https://doi.org/10.32942/X24891
Astrobiology