Active Prokaryotic And Eukaryotic Viral Ecology Across Spatial Scale In A Deep-sea Brine Pool
Deep-sea brine pools represent rare, extreme environments that focus biodiversity at bathyal to abyssal depths. Despite their small size and distribution, brine pools represent important ecosystems to study because they provide unique insight into the limits of life on Earth, and by analogy, the plausibility of life beyond it.
A distinguishing feature of many brine pools is the presence of thick benthic microbial mats which develop at the brine-seawater interface. While these bacterial and archaeal communities have received moderate attention, little is known about the viral communities and their interactions with host populations in these environments.
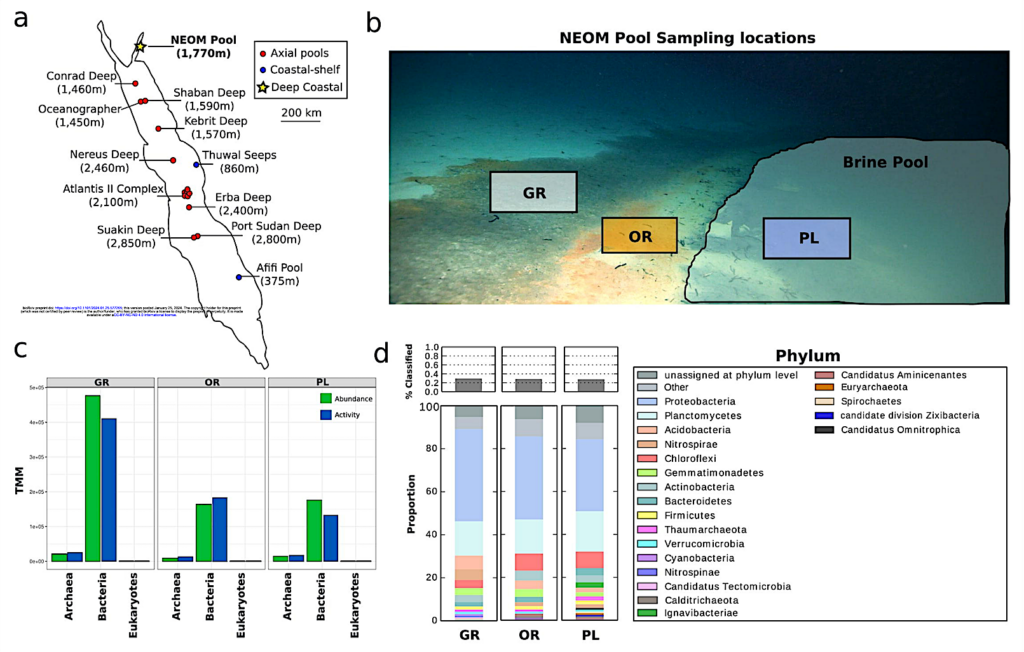
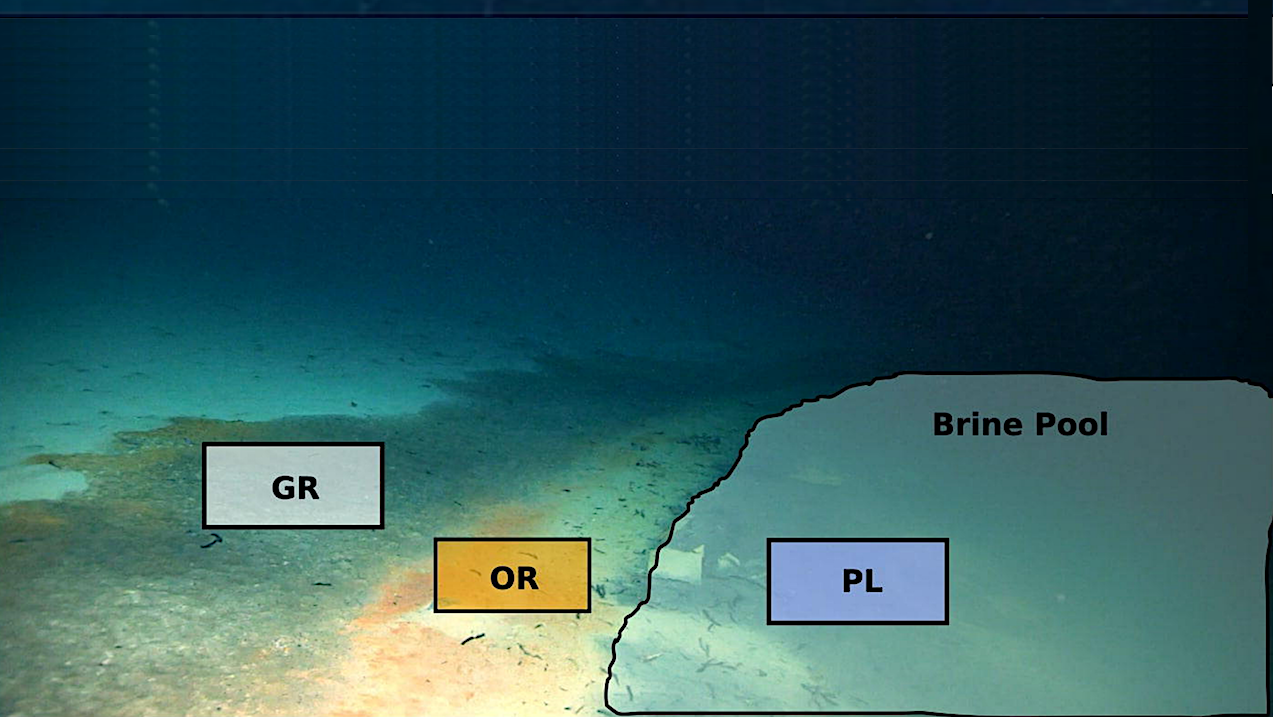
Sampling location and community composition. (a) A map of all discovered Red Sea brine pools, split into three categories based on location and depth. (b) A picture taken from the remotely operated vehicle (ROV) of the sampling locations showing the gray microbial mat (GR), orange mat (OR), and pool zones (PL). (c) Abundance and activity of different kingdoms of life based on normalized metagenomic (abundance) and metatranscriptomic (activity) reads (TMM) mapped to contigs from each kingdom identified using Tiara. (d) Relative abundance of different bacterial and archaeal phyla at each zone. These proportions were determined using kaiju to assign taxonomy to trimmed metagenomic reads. — biorxiv.org
To bridge this knowledge gap, we leveraged metagenomic and metatranscriptomic data from three distinct zones within the NEOM brine pool system (Gulf of Aqaba) to gain insights into the active viral ecology around the pools.
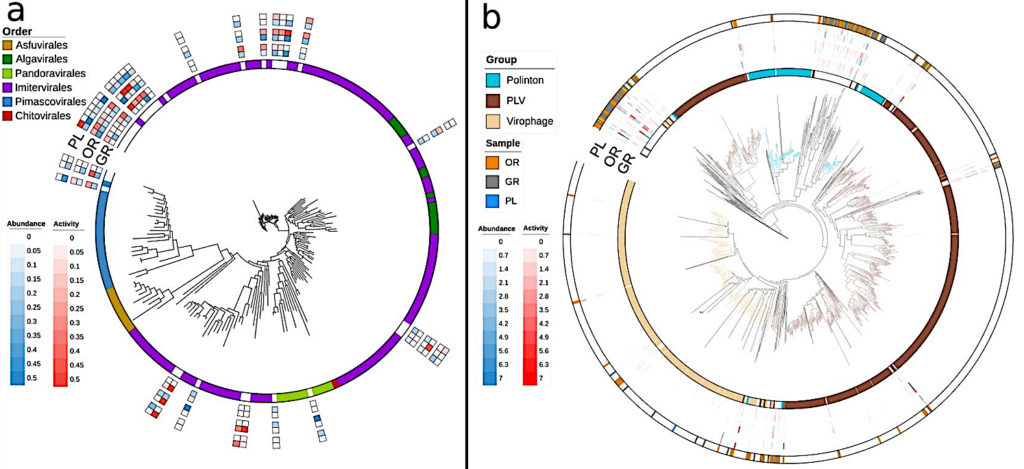
Here, we report a remarkable diversity and activity of viruses of all nucleic acid types and genome sizes that infect prokaryotic and eukaryotic hosts in this environment. These include giant viruses (phylum: Nucleocytoviricota), RNA viruses, jumbo phages, and polinton-like viruses (PLVs).
Predicted and potential hosts for prokaryotic and eukaryotic viruses. (a) Virushost predictions for prokaryotic viruses were done using iPHOP for all zones. A total of 36 host predictions were made across zones and band thickness represents more hosts from a given bacterial class or zone. (b) The potential eukaryotic host pool was created through mapping reads to confirmed eukaryotic contigs obtained from Tiara. These contigs were classified to the phylum level using CAT and proportions represent RPKM values for each zone. — biorxiv.org
Many of these appeared to form distinct clades showing the possibility of untapped viral diversity in the brine pool ecosystem. Zone-specific differences in viral community composition and infection strategy were also observed with lysogenic phages seeming to dominate the bacterial mat further away from the pool’s center.
Through host matching, viruses infecting metabolically important bacteria and archaea were observed – including a linkage between a jumbo phage and a key manganese-oxidizing and arsenic-metabolizing bacterium. Our findings shed light on the role of viruses in modulating the brine pool microbial community dynamics and biogeochemistry through revealing novel viral diversity, host-virus associations, and spatial-scale heterogeneity in viral dynamics in these extreme environments.
These results will provide crucial foundation for further investigation into the adaptations of viruses and their microbial hosts in extreme habitats in the marine ecosystem.
Benjamin Minch, Morgan Chakraborty, Sam Purkis, Mattie Rodrigue, Mohammad Moniruzzaman
Active prokaryotic and eukaryotic viral ecology across spatial scale in a deep-sea brine pool, biorxiv.org
doi: https://doi.org/10.1101/2024.01.25.577265
Astrobiology, Genomics,