A Conserved Ribosomal Protein Has Entirely Dissimilar Structures In Different Organisms
Ribosomes from different species can markedly differ in their composition by including dozens of ribosomal proteins that are unique to specific lineages but absent in others. However, it remains unknown how ribosomes acquire and specialize new proteins throughout evolution.
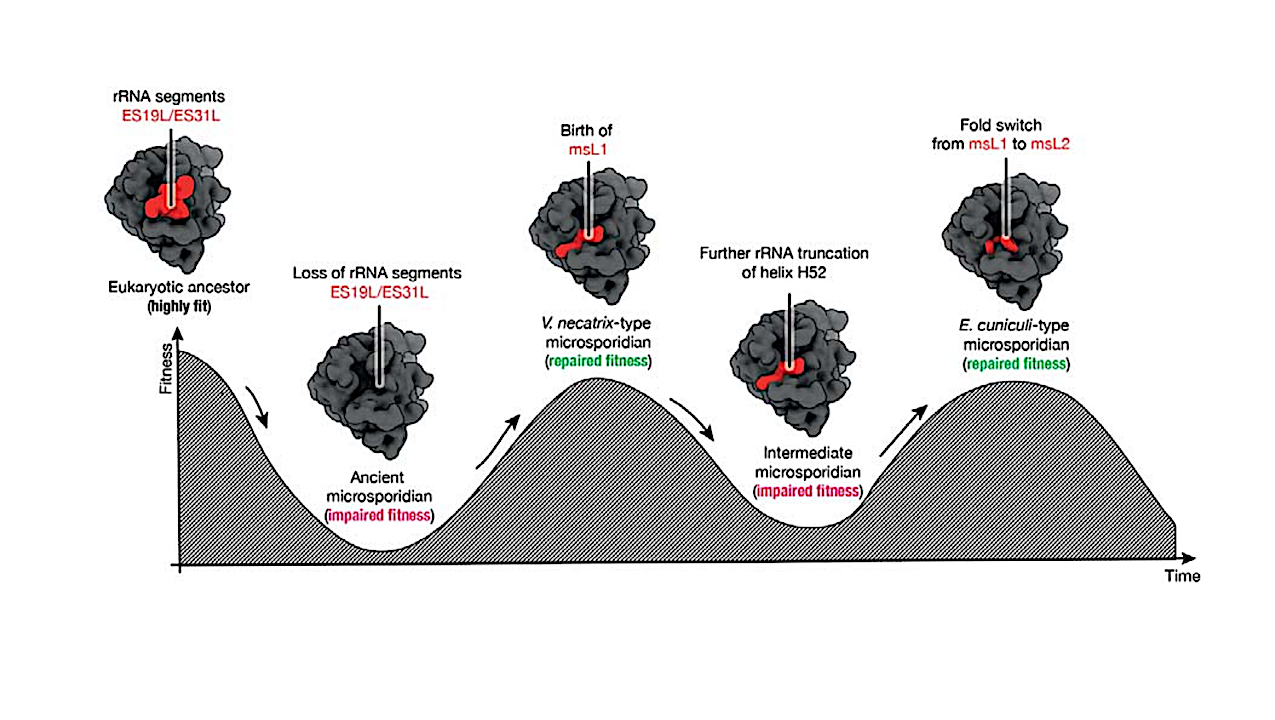
Here, to help answer this question, we describe the evolution of the ribosomal protein msL1/msL2 that was recently found in ribosomes from the parasitic microorganism clade, microsporidia. We first show that this protein has a conserved location in the ribosome but entirely dissimilar structures in different organisms: in each of the analyzed species, msL1/msL2 exhibits an altered secondary structure, an inverted orientation of the N- and C-termini on the ribosomal binding surface, and a completely transformed three-dimensional fold. We then show that this evolutionary fold switching is likely caused by changes in the msL1/msL2-binding site in the ribosome; specifically, by variations in microsporidian rRNA.
These observations allow us to infer an evolutionary scenario in which a small, positively-charged, de novo-born unfolded protein was first captured by rRNA to become part of the ribosome and subsequently underwent complete fold switching to optimize its binding to its evolving ribosomal binding site. Overall, our work provides a striking example that will help us better understand the origin and evolution of new protein components of complex molecular assemblies – thereby enhancing our ability to engineer biological molecules, identify protein homologs, and peer into the history of life on Earth.
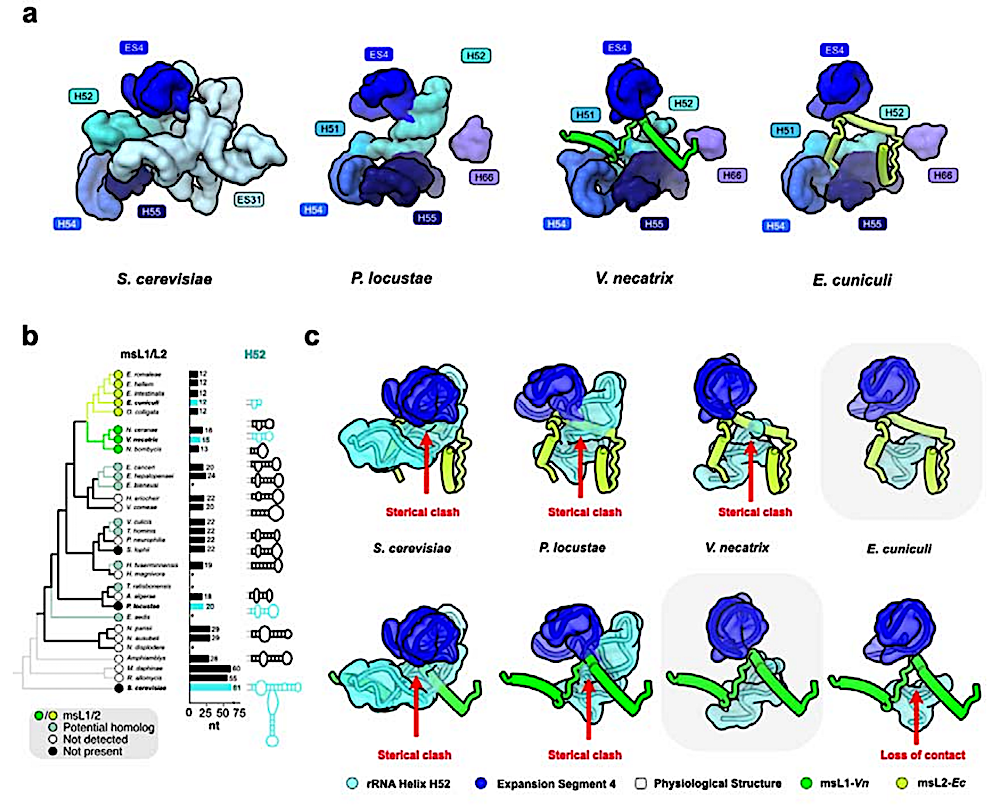
The msL1/msL2 fold change is likely caused by rRNA truncations. a) The structures compare ribosomal binding pockets of msL1-Vn and msL2-Ec and their corresponding ribosome segments in S. cerevisiae and P. locustae. b) A ribosomal protein-based phylogenetic tree is shown with detectable msL1/msL2 homologs indicated. For species with available 5.8S-25S rRNA sequences, the length and predicted secondary structure of the helix H52 are shown next to their corresponding species. Species with missing or incomplete sequences for 5.8S-25S rRNA are highlighted by asterisks, and species with experimentally defined ribosome structures are highlighted in cyan. c) The structures of msL1-Vn and msL2-Ec docked into the ribosomal environment of S. cerevisiae, P. locustae, V. necatrix and E. cuniculi ribosomes to illustrate potential steric clashes or loss of ribosome-binding surface in one species relative to each other. — biorxiv.org
Léon Schierholz, Charlotte R. Brown, Karla Helena-Bueno, Vladimir N. Uversky, Robert H. Hirt, Jonas Barandun, Sergey V. Melnikov
doi: https://doi.org/10.1101/2022.09.21.508910
Now published in Molecular Biology and Evolution doi: 10.1093/molbev/msad254
A conserved ribosomal protein has entirely dissimilar structures in different organisms
Astrobiology, genomics, evolution