Developing An AI Method Of Identifying Life On Other Worlds

Humankind is looking for life on other planets, but how will we recognise it when we see it? Now a group of US scientists have developed an artificial-intelligence-based system which gives 90% accuracy in discovering signs of life.
The work was presented to scientists for the first time at the Goldschmidt Geochemistry Conference in Lyon on Friday 14th July, where it received a positive reception from others working in the field. The details have now been published in the peer-reviewed journal PNAS (see notes for details).
Lead researcher Professor Robert Hazen, of the Carnegie Institution’s Geophysical Laboratory and George Mason University. said “This is a significant advance in our abilities to recognise biochemical signs of life on other worlds. It opens the way to using smart sensors on unmanned spaceships to search for signs of life”.
Since the early 1950’s scientists have known that given the right conditions, mixing simple chemicals can form some of the more complex molecules required for life, such as amino acids. Since then, many more of the components necessary for life, such as the nucleotides needed to make DNA, have been detected in space. But how do we know if these are of biological origin, or if they are made by another abiotic process over time. Without knowing that, we don’t know if we have detected life.
Bob Hazen said “We are asking a fundamental question; Is there something fundamentally different about the chemistry of life compared to the chemistry of the inanimate world? Are there “chemical rules of life” that influence the diversity and distribution of biomolecules? Can we deduce those rules and use them to guide our efforts to model life’s origins or to detect subtle signs of life on other worlds? We found that there is.
From an evolutionary point of view, life is not an easy thing to sustain, and so there are certain pathways which work and certain which don’t. Our analysis does not rely on absolute identification of a compound but determines biological/non-biological origins by looking at the compound in relation to the sample context”.
What they did
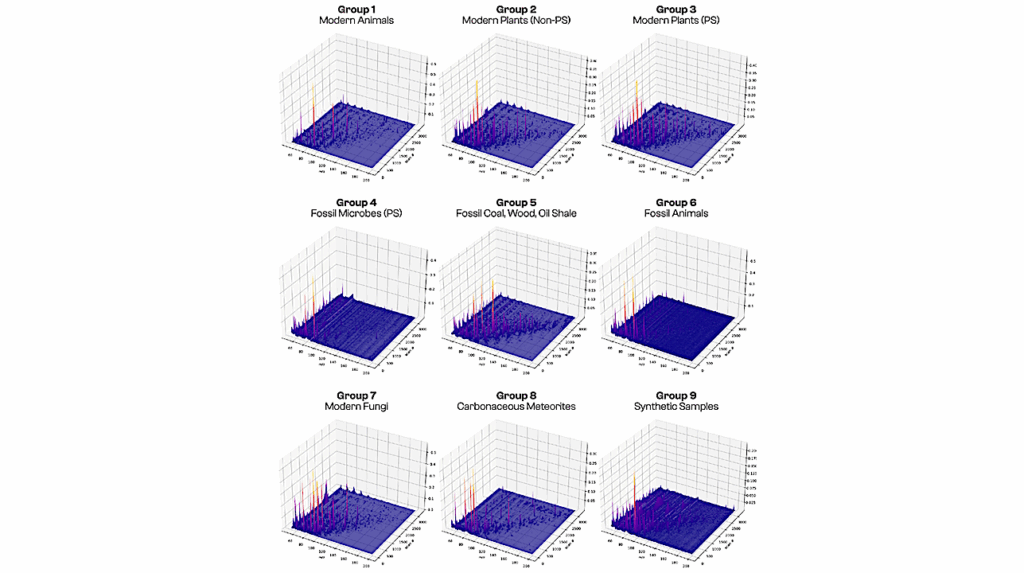
The scientists employed NASA flight-tested* pyrolysis gas-chromatography mass-spectrometry (GCMS) methods to analyse 134 varied carbon-rich samples from living cells, age-degraded samples, geologically processed fossil fuels, Carbon-rich meteorites, and laboratory-synthesized organic compounds and mixtures. 59 of these were of biological origin (biotic), such as a grain of rice, a human hair, crude oil, etc. 75 were of non-biological origin (abiotic), such as lab-synthesised compounds like amino acids, or samples from carbon-rich meteorites. The samples were first heated in an oxygen-free environment, which causes the samples to break down (a process known as pyrolysis). The treated samples were then analysed in a GC-MS, an analytical device which separates the mixture into its component parts, and then identifies them. Using a suite of machine-learning methods, three-dimensional (time/intensity/mass) data from each abiotic or biotic sample were employed as training or testing subsets, which resulted in a model that can predict the abiotic or biotic nature of the sample with greater than 90 percent accuracy.
The first presentation and feedback from other scientists
Professor Hazen presented the work for the first time to scientists at the Goldschmidt geochemistry conference in Lyon, France, on 14th July, as part of a session looking at geobiology of life on Earth and other planetary systems (https://conf.goldschmidt.info/goldschmidt/2023/meetingapp.cgi/Session/5290).
In response to questions from the audience, Professor Hazen confirmed that “The team will be able to expand the range of biosignatures, to detect extraterrestrial life, which may be fundamentally different to life on Earth”.
Session co-chairs, Anastasia Yanchilina (Impossible Sensing, St Louis), and Fabian Gäb (University of Bonn) noted that that the in-person feedback from the attending scientists was lively and positive.
Dr Yanchilina said “The session as a whole went well, and this talk was one of the cherries on the cake. This moves us closer to recognising life when we find it”.
What it means
Professor Hazen continued “There are some interesting and deep implications which flow from this work. First, we can apply these methods to ancient samples from Earth and Mars, to find out if they were once alive. This is obviously important for looking at whether there was life on Mars, but it can also help us analyse very ancient samples from Earth, to help us understand when life first began.
It also means that at a deep level, biochemistry and non-biological chemistry are somehow different. This probably also means that we may be able to tell a lifeform from another planet, from another biosphere, from the ones we know on Earth. This means that if we find life elsewhere, we can tell if life on Earth and other planets came from a common origin (panspermia), or whether they would have come from different origins.
What really astonished us was that we trained our machine-learning method on only two attributes–biotic or abiotic–but the method discovered 3 distinct populations–abiotic, living biotic, and fossil biotic – in other words, it could tell fossil samples from more recent biological samples. This surprising finding gives us optimism that other attributes such as photosynthetic life or eukaryotes (cells with a nucleus) might also be distinguished.
In summary, this study is just the beginning of what may become a widely useful approach to teasing out information from enigmatic organic mixtures”.
Commenting, Professor Emmanuelle Javaux (Head of Early Life Traces and Evolution-Astrobiology lab, Director of Research unit Astrobiology, University of Liège, Belgium) said:
“I think this new study is very exciting. It is a new avenue of research to explore as it appears to discriminate abiotic from biotic organic matter based on its molecular complexity and could potentially be a fantastic tool for astrobiology missions. It would also be very interesting to test this new method on some of the oldest putative and debated traces of Earth life as well as on modern and fossil organisms from the three domains of life! this might help to solve some hot debates in our community”.
This is an independent comment, Professor Javaux was not involved in this work.
*This PY-GC-MS system has been flight-tested on the Viking mission and the Curiosity Rover, both missions to Mars.
NOTES
This work was presented at the Goldschmidt conference. A paper on the work will be published in the peer-reviewed journal PNAS (Proceedings of the National Academy of Sciences) on 25th September A robust agnostic molecular biosignature based on machine learning, PNAS
Astrobiology