Exoplanet Characterization With NASA’s Habitable Worlds Observatory

Executive Summary: The Habitable Worlds Observatory (HWO) is the first astrophysics flagship mission with a key cross-divisional astrobiology science goal of searching for signs of life on rocky planets beyond our solar system.
The Living Worlds Working Group under the Science, Technology, and Architecture Review Team (START) was charged with investigating how HWO could characterize potentially habitable exoplanets orbiting stars in the solar neighborhood, search for signs of life, and interpret potential biosignatures within a false positive and false negative framework.
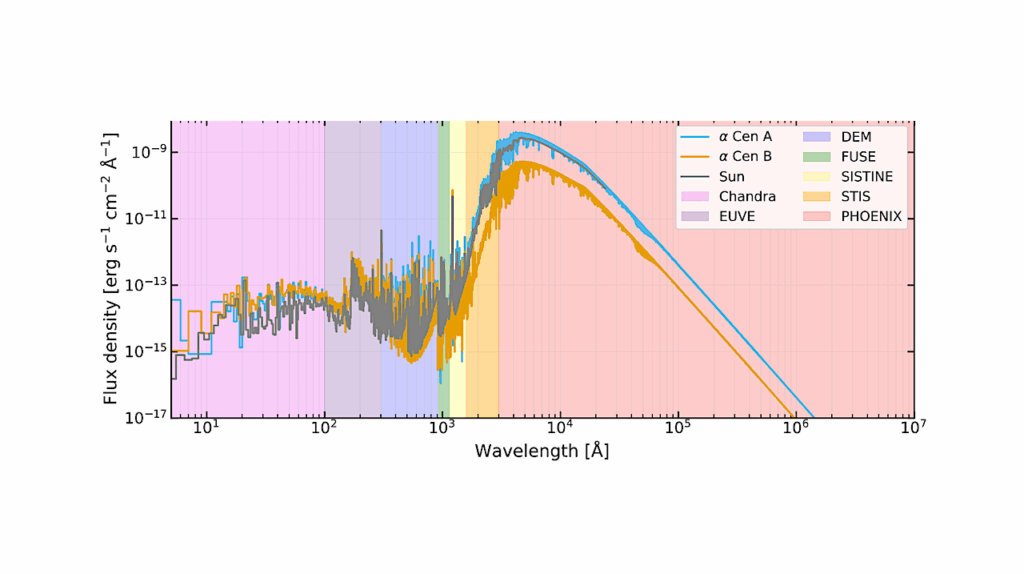
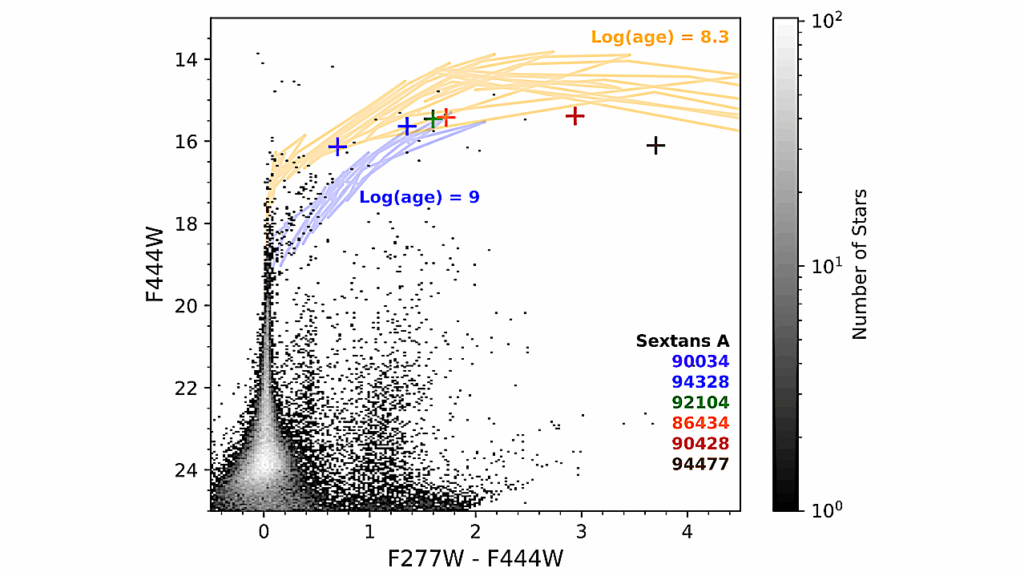
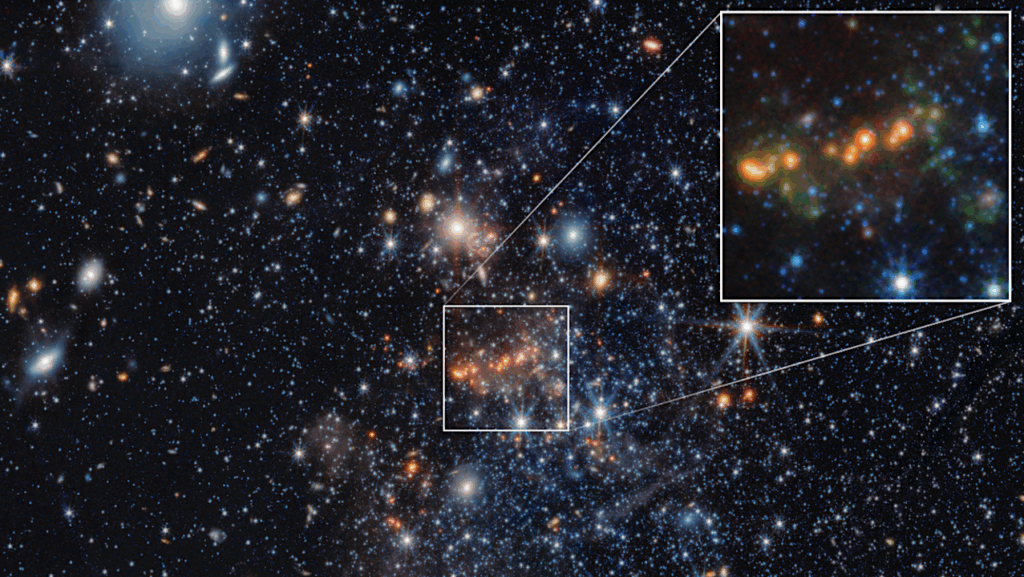
In particular, we focused on (1) identifying biosignatures that have spectral features in the UV-Vis-NIR wavelength range and defining their measurement requirements, (2) determining additional information needed from the planet and planet system to interpret biosignatures and assess the likelihood of false positives, and (3) assembling current knowledge of likely HWO target stars and identify which properties of host stars and systems are most critical to know in advance of HWO.
The Living Worlds atmospheric biosignatures science case is considered one of the key drivers in the design of the observatory. An additional 10 astrobiology science cases were developed that collectively revealed key research gaps and needs required to fully explore the observatory parameter space and perform science return analyses. Investment in these research gaps will require coordination across the Science Mission Directorate and fall under the purview of the new Division-spanning astrobiology strategy.
NASA Decadal Astrobiology Research and Exploration Strategy (NASA-DARES 2025) White Paper — Habitable Worlds Observatory Living Worlds Science Cases: Research Gaps and Needs
Niki Parenteau, Giada Arney, Eleanora Alei, Ruslan Belikov, Svetlana, Berdyugina, Dawn Cardace, Ligia F. Coelho, Kevin Fogarty, Kenneth Gordon, Jonathan Grone, Natalie Hinkel, Nancy Kiang, Ravi Kopparapu, Joshua Krissansen-Totton, Emilie LaFleche, Jacob Lustig-Yaeger, Eric Mamajek, Avi Mandell, Taro Matsuo, Connor Metz, Mark Moussa, Stephanie Olson, Lucas Patty, Bill Philpot, Sukrit Ranjan, Edward Schwieterman, Clara Sousa-Silva, Anna Grace Ulses, Sara Walker, Daniel Whitt
Comments: This white paper was submitted to the NASA Decadal Astrobiology Research and Exploration Strategy (NASA-DARES 2025) RFI. 5 pages, 1 figure
Subjects: Instrumentation and Methods for Astrophysics (astro-ph.IM); Earth and Planetary Astrophysics (astro-ph.EP)
Cite as: arXiv:2601.06386 [astro-ph.IM] (or arXiv:2601.06386v1 [astro-ph.IM] for this version)
https://doi.org/10.48550/arXiv.2601.06386
Focus to learn more
Submission history
From: Niki Parenteau
[v1] Sat, 10 Jan 2026 02:14:07 UTC (1,239 KB)
https://arxiv.org/abs/2601.06386
Astrobiology,