Planetary Analog Sites In The Indian Subcontinent And The Indian Ocean: Underexplored Environments Suited For Astrobiological And Space Research
Astrobiology seeks to understand the origin, evolution, distribution, and future of life in the Universe, focusing on habitability beyond Earth.
Due to the high cost and complexity of space missions, studying planetary analog sites on Earth is essential for supporting and de-risking future exploration. These analog sites are extreme terrestrial environments that mirror environmental, geological, geochemical, or biological conditions on other planetary bodies.
Investigating how life persists in these settings advances knowledge of extraterrestrial habitability and enables realistic testing of life-detection instruments. This review presents the first comprehensive synthesis of more than 50 planetary analog field sites across the Indian subcontinent and Indian Ocean region.
We identify 2 geological regions with active astrobiological research, 4 requiring targeted geochemical and geomicrobiological surveys, and 5 with high planetary relevance but minimal study. We assess how these sites fill gaps in global astrobiological research and evaluate their readiness for future investigations.
The sites include high-altitude cryospheric settings such as Himalayan glaciers and permafrost, analogues to Martian and lunar environments; saline-alkaline lakes like Sambhar Lake, comparable to Martian paleolakes; intrabasaltic bole beds in the Deccan Traps, relevant to phyllosilicate formation on Mars; subsurface caves and mines, analogous to lunar lava tubes; and hydrothermal vent systems along the Central and Southwest Indian Ridges, relevant to icy ocean worlds. Comparing these sites to global analogues reveals that, although most are not yet fully characterized, several offer unique environmental combinations.
As deep-space missions prepare to search for life beyond Earth, a geographically broader set of analog sites is critical. Highlighting the diversity and scientific value of these under-characterized regions in South Asia and their marine periphery, this review provides a foundation for characterizing previously overlooked planetary analog sites. These sites expand the global analogue parameter space and offer underutilized natural laboratories for planetary habitability and biosignature research.

Planetary analog environments across the Indian subcontinent and the Indian ocean basins that represent planetary bodies of astrobiological interest. — Frontiers via eartharxiv.org
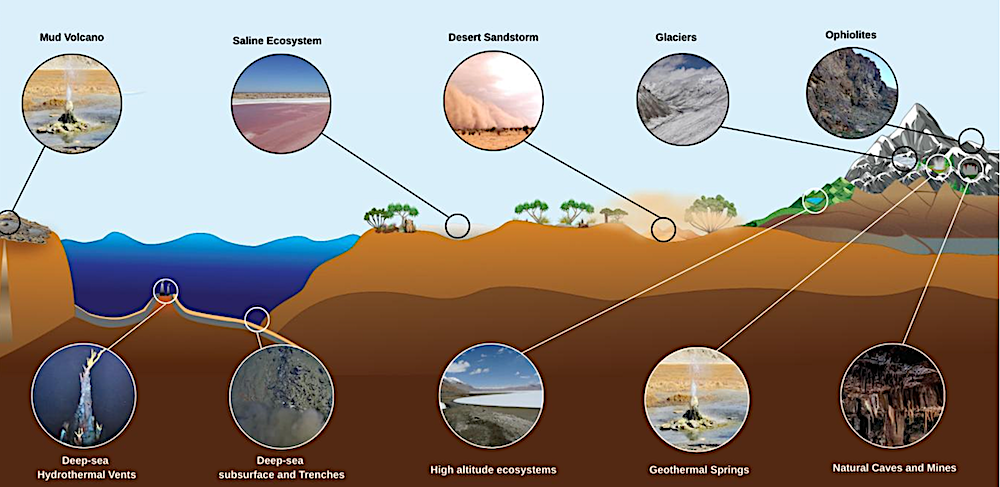
Schematic representation of extreme environments across the Indian subcontinent and the Indian Ocean that serve as planetary analogues. Terrestrial ecosystems include mud volcanoes, saline habitats, desert sandstorms, glaciers, ophiolites, geothermal springs, high-altitude lakes, and natural caves and mines. Marine ecosystems of the Indian Ocean remain relatively underexplored, with documented sites including deep-sea hydrothermal vents and subsurface trenches. This figure is adapted and inspired by Merino et al. (2019) — Frontiers via eartharxiv.org
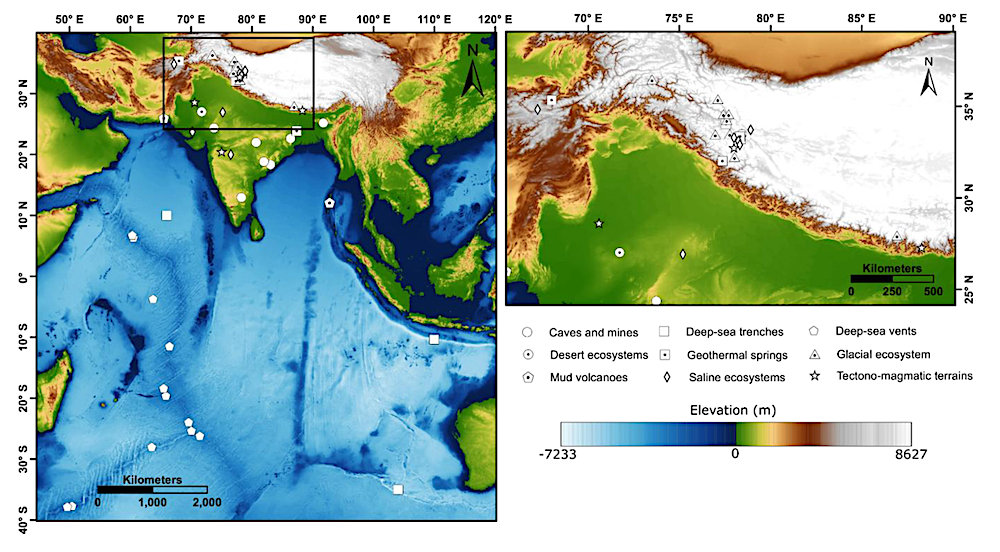
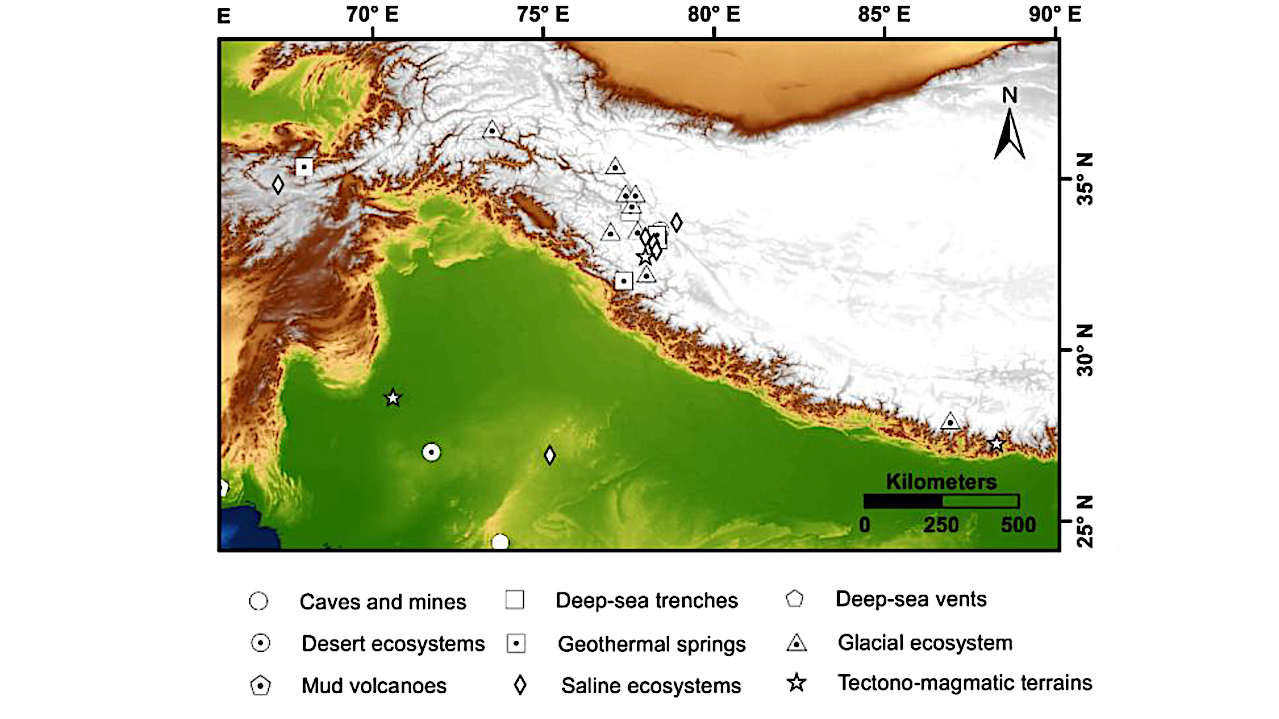
Geographic distribution of extreme environments, that can serve as planetary analogues, overlaid on a hypsometric map of the Indian subcontinent and adjoining Indian Ocean. The left panel illustrates the regional distribution of terrestrial and marine sites across diverse ecosystems, while the right panel provides a detailed view of the Himalaya and Tibetan Plateau, highlighting clusters of high-altitude and glacial environments. The underlying topography and bathymetry emphasize the environmental gradients that shape the diversity of analogue sites. Geological base map compiled from GEBCO Compilation Group (2025). — Frontiers via eartharxiv.org
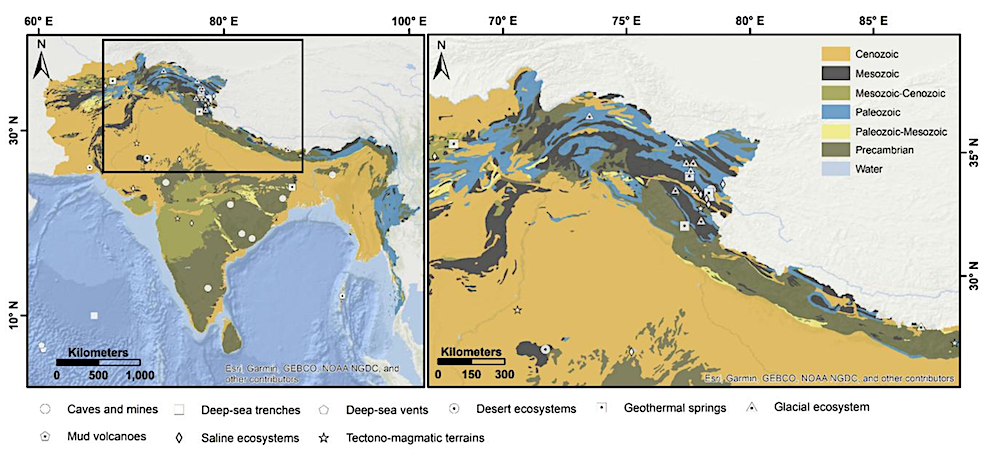
Geographic distribution of extreme environments, that can serve as planetary analogues, overlaid on a generalized geological map of the bed rock of the Indian subcontinent. The underlying geological framework (Cenozoic, Mesozoic, Paleozoic, and Precambrian units) highlights the lithological diversity associated with these analogue sites. Geological base map compiled from Wandrey, C.J., 1998, Geologic map of South Asia (geo8ag). — Frontiers via eartharxiv.org
- Planetary analog sites in the Indian subcontinent and the Indian Ocean: Underexplored environments suited for astrobiological and space research, eartharxiv.org
- Planetary analog sites in the Indian subcontinent and the Indian Ocean: Underexplored environments suited for astrobiological and space research, Frontiers
Astrobiology,