Emergence Of Diverse Ligase Reactivities From A Single RNA Evolution Experiment
RNA-catalyzed assembly of RNA strands was one of the most essential enzymatic functions in a primordial RNA-based biology.
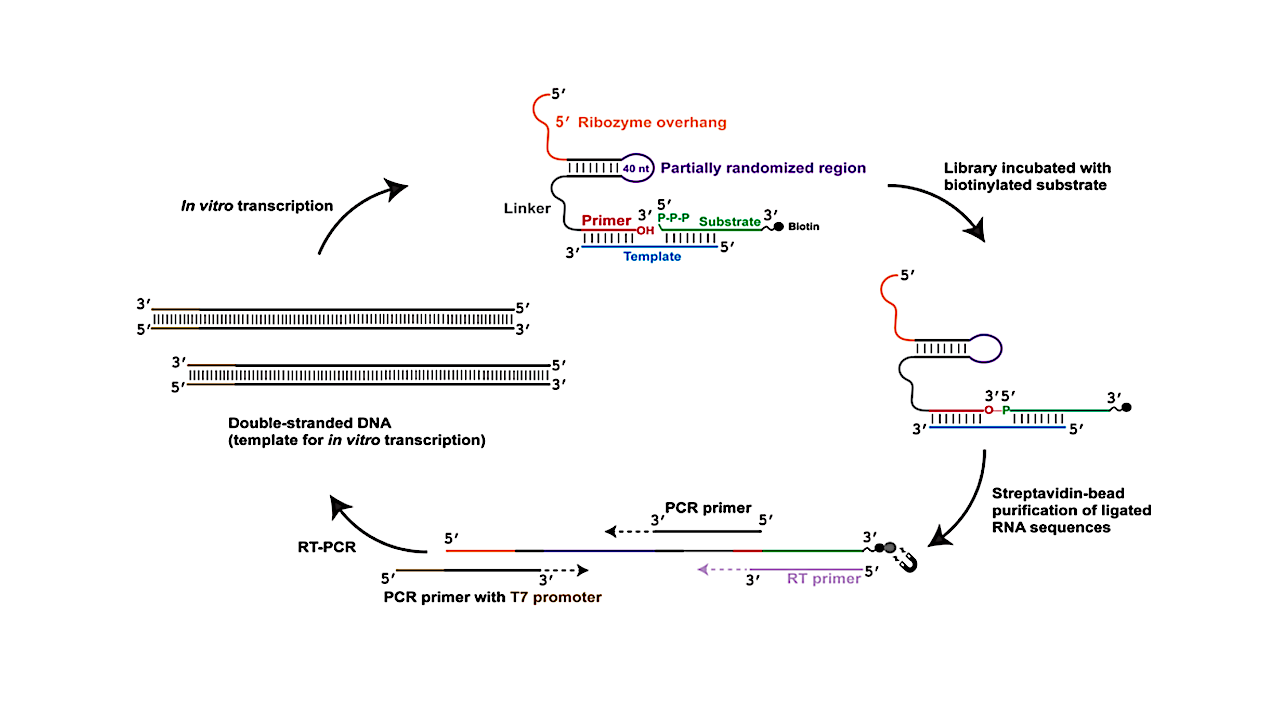
To explore the scope of RNA-catalyzed RNA assembly, we used directed evolution to re-engineer a ligase ribozyme that originally used 5′-phosphorimidazolide RNA substrates into a ribozyme that ligates RNA substrates carrying the biologically relevant 5′-triphosphate group.
Unexpectedly, analysis of the low-abundance regime of the selected RNA population, representing ∼0.1% of the population, revealed four distinct types of ligase ribozymes, indicating that multiple ribozymes had emerged from a single experiment, despite the stringent selection for the desired ‘triphosphate ligase’.
The first ligase type exhibits strict specificity for 5′-phosphorimidazolide substrates, even though these substrates were never presented during selection. The second and third ligase types catalyze two different branching reactions, each involving the 5′-triphosphate groups on the ribozyme and a different internal hydroxyl group on the substrate.
These reactions resemble ‘branching’ reactions catalyzed by naturally occurring ribozymes, including the spliceosome, even though these ribozymes emerged from a synthetic RNA library with no relation to biological RNA catalysts. The fourth ligase type mediates a reaction between its 5′-triphosphate and the substrate’s terminal 2′-OH, but only when the substrate carries a 3′-phosphate.
Remarkably, a single point mutation toggles this ribozyme between linear ligation and branching, functioning as a unique reactivity switch. Such a switch provides a mechanism for RNA catalysts to acquire new reactivities with minimal mutational perturbation, and therefore, has interesting evolutionary implications for primordial biocatalysis.
More generally, the emergence of such catalytic diversity from a single evolution experiment under highly constrained selection pressures highlights the catalytic flexibility and evolutionary potential of RNA. These findings strengthen the plausibility that an RNA World could have supported a wide range of chemistries essential for the emergence of life.
Emergence of diverse ligase reactivities from a single RNA evolution experiment, biorxiv.org
Astrobiology