A New Evolutionary Scenario Marked By Massive Gene Loss And Expansion
Evolution is traditionally associated with a process of increasing complexity and gaining new genes.
However, the explosion of the genomic era shows that gene loss and simplification is a much more frequent process in the evolution of species than previously thought, and may favour new biological adaptations that facilitate the survival of living organisms. This evolutionary driver, which seems counter-intuitive — “less is more” in genetic terms — now reveals a surprising dimension that responds to the new evolutionary concept of “less, but more”, i.e. the phenomenon of massive gene losses followed by large expansions through gene duplications.
This is one of the main conclusions of an article published in the journal Molecular Biology and Evolution, led by a team from the Genetics Section of the Faculty of Biology and the Institute for Research on Biodiversity (IRBio) of the University of Barcelona, in which teams from the Okinawa Institute of Science and Technology (OIST) have also participated. The paper identifies new evolutionary patterns, and it outlines a new scenario, marked by the enormous potential for genetic change and evolutionary adaptation driven by large-scale gene loss and duplication in living organisms.
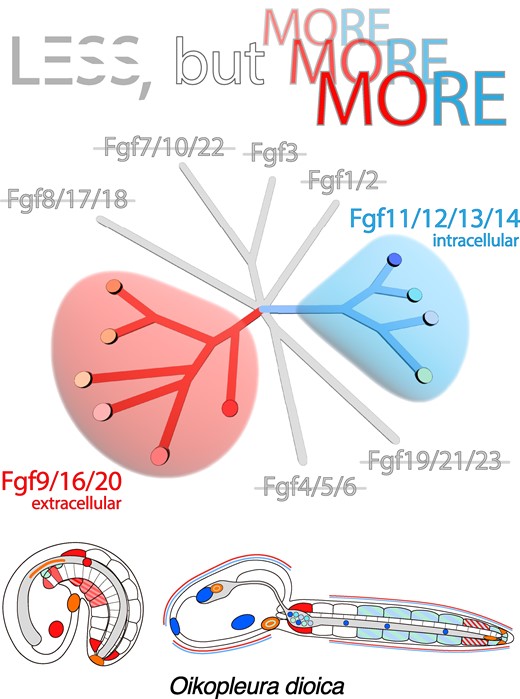
Graphical Abstract — Animals
Less, but more: a new evolutionary scenario
There are still many questions about the impact of gene loss on the diversification of species and the emergence of evolutionary innovations in the planetary tree of life. “Gene loss is a widespread mechanism throughout the biological scale and represents an evolutionary driving force that can generate genetic variability and also biological adaptations, and this has traditionally been known as the ‘less is more’ hypothesis”, says Cristian Cañestro, leader of the study and member of the UB’s consolidated research group on Evolution and Development (Evo-Devo) of the UB’s Department of Genetics, Microbiology and Statistics.
Now, the new paper describes a new evolutionary framework called “less, but more”, which extends the previous model in terms of the importance of gene loss as an evolutionary driving force.
This study, which is part of Gaspar Sánchez-Serna’s doctoral thesis, focuses on the study of the genome of the Oikopleura dioica species, a swimming organism of the marine zooplankton that belongs to the tunicates — a sister group of vertebrates — and is phylogenetically linked to evolutionary history. In this study model — a free-living tunicate or appendicularian — the team reconstructed the evolutionary history of fibroblast growth factor (FGF) gene families, which are critical in the developmental process of organisms.
“The findings suggest that the process of gene loss reduced the number of FGF growth factor gene families from eight to just two, which are the Fgf9/16/20 and Fgf11/12/13/14 families. These surviving subfamilies have doubled over the course of evolution to generate a total of ten genes in appendicularians”, explains Sánchez-Serna, first author of the paper. “In particular, Fgf9/16/20 and Fgf11/12/13/14 may represent a minimal set of subfamilies that have conserved secretory and intracellular functions, respectively, and reveal important information about the evolution of the FGF system”, he continues.
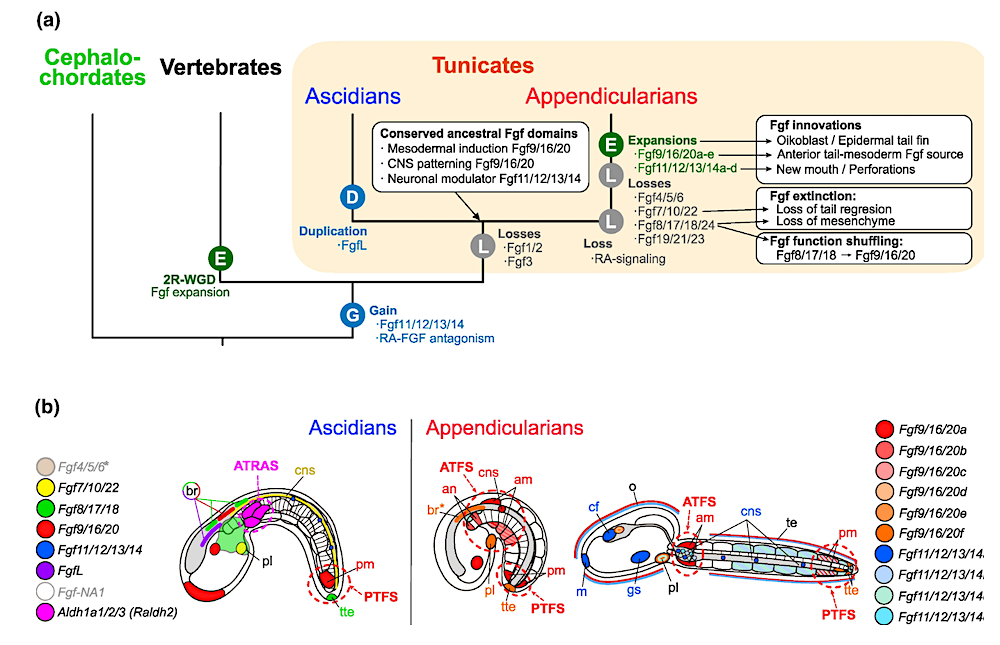
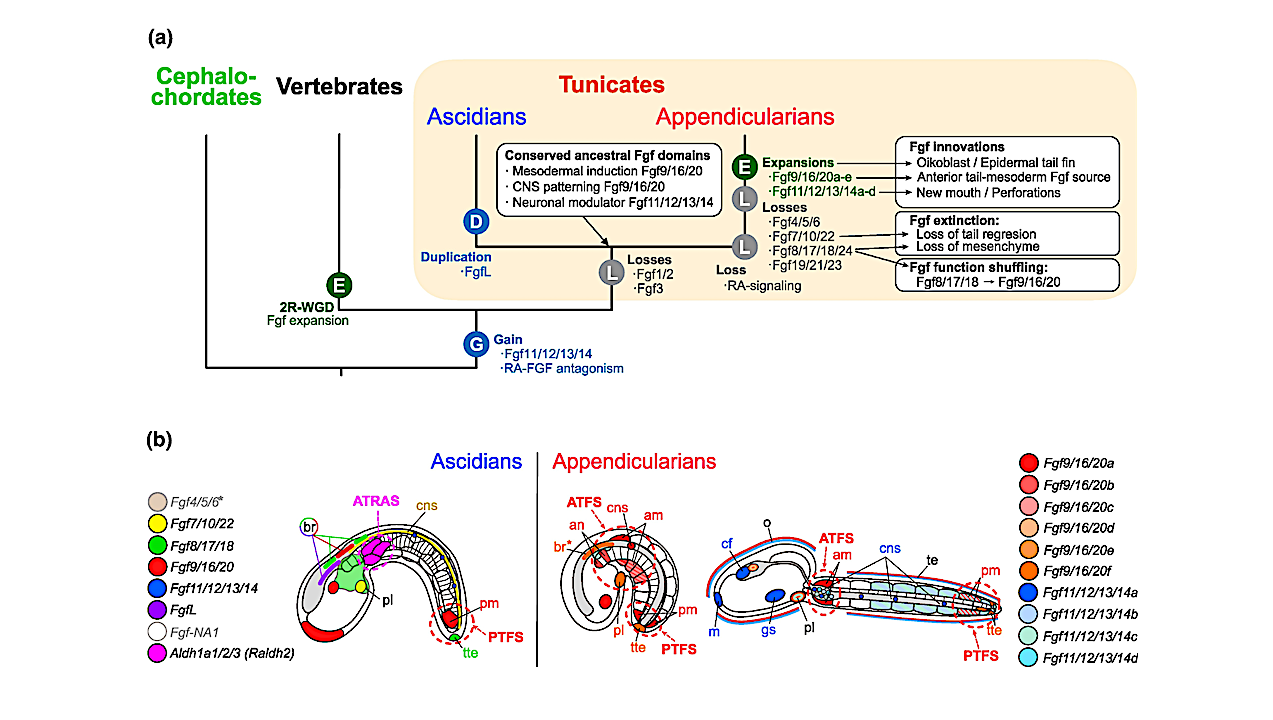
Evolutionary scenario of Fgf subfamilies in chordates. a) Evolutionary tree of chordate subphyla indicating main events of losses (L), gains (G), duplications (D), or expansions by burst of duplications (E), as well as main associated patterns of conservation, innovation, function shuffling and extinction of Fgf expression domains. 2R-WGD: two rounds of whole-genome duplication. b) Comparative schematic representation of the main expression domains of Fgf subfamily members between ascidians and appendicularians (tailbud stage in the left, and hatchling in the right). Distinct colors are assigned to each Fgf as indicated in the figure legend. FgfNA1 expression has not been described in ascidians, and Fgf4/5/6* has been detected maternally and widely throughout development with no obvious tissue-specific domains (Imai et al. 2004). In appendicularians, the expression of Fgf9/16/20 paralogs is the most abundant in tailbud stages, and the expression of Fgf11/12/13/14 paralogs is more obvious at hatchling stages. Asterisk (in br*) denotes that expression was found in a slightly earlier stage than the one represented in the figure. Ascidians show a characteristic PTFS (Posterior-Tail FGF Source) and ATRAS (Anterior-Tail RA Source), while in appendicularians the loss of RA signaling (Cañestro and Postlethwait 2007; Martí-Solans et al. 2016) might be related to the innovation of an ATFS (Anterior-Tail FGF Source), considering that the conserved RA-FGF antagonistic action emerged at the base of olfactores (Pasini et al. 2012; Bertrand et al. 2015). Abbreviations: an, anterior notochord; am, anterior muscle; br, brain; cf, ciliary funnel; cns, central nervous system; gs, gill slit; m, mouth; o, oikoblastic epithilium; pl, placode; pm, posterior muscle; te, tail epidermis; tte, terminal tail epidermis. — Animals
From sessile life to active swimming
The study provides a new perspective on the evolution of FGF subfamilies in the chordate group, with massive losses and duplications of ancestral gene families originating at the base of the appendicular lineage after they diverged from the ascidians. All these changes have contributed to morphological divergence between different species of free-living tunicates, such as O. dioica.
“Our study presents a new hypothesis on how FGF gene losses and duplications may be related to developmental changes. We are talking about evolutionary innovations — changes in morphology and body plan, etc. — that drove the evolution from the ascidian-like sessile lifestyle to free-living, actively swimming forms such as appendicularians”, says Sánchez-Serna.
The study also identifies differences in the structure of the FGF genes of O. dioica from different parts of the world, providing the first molecular evidence of how these rapidly evolving populations are becoming cryptic species (i.e. consisting of organisms very similar in morphology and genome that have hitherto been classified in the same species).
The “less, but more” evolutionary model “helps us to understand how sometimes losing opens up new possibilities for subsequent gains and, therefore, losses are necessary to favour the evolutionary origin of new adaptations”, concludes Cristian Cañestro.
Less, but more: new insights from appendicularians on chordate Fgf evolution and the divergence of tunicate lifestyles, Animals (open access)
Astrobiology,