Tricorder Tech: New Method Tracks The ‘Learning Curve’ Of AI To Decode Complex Genomic Data
Editor’s note: If we aspire to mount expeditions to new worlds and then embrace the task of characterizing and quantifying whatever life forms we find, the ability to map and understand whatever metabolic and genomic systems are in operation is important. Not only do we need to know how alien biota function, but also how they evolved – what differences and similarities they may have with the origin and evolution of life on Earth. Increasing in situ capabilities like this can allow much more preliminary analysis to be done on site – or back on Earth.
As we begin to expand our search for life to other worlds we are going to need to be economical interms of the equipment we send and how we reality new knowledge back to Earth. Sample return missions are difficult even when worlds are close to one another. Doing in situ examination and documentation is going to be very important as we explore other worlds. Not only does it reduce the logistics of sending things back home but it allows data to be sent back at the speed of light. It also allows the astronaut/droid teams to engage in empirical exploration – learning from what they found so as to refine and perfect their continued searching.
Introducing Annotatability—a powerful new framework to address a major challenge in biological research by examining how artificial neural networks learn to label genomic data. Genomic datasets often contain vast amounts of annotated samples, but many of these samples are annotated either incorrectly or ambiguously.
Borrowing from recent advances in the fields of natural language processing and computer vision, the team used artificial neural networks (ANNs) in a non-conventional way: instead of merely using the ANNs to make predictions, the group inspected the difficulty with which they learned to label different biological samples.
Somewhat similarly to assessing why students find some examples harder than others, the team then leveraged this unique source of information to identify mismatches in cell annotations, improve data interpretation, and uncover key cellular pathways linked to development and disease.
Annotatability provides a more accurate method for analyzing genomic data on single cells, offering significant potential for advancing biological research, and in the longer term, improving disease diagnosis and treatment.
A new study led by Jonathan Karin, Reshef Mintz, Dr. Barak Raveh and Dr. Mor Nitzan from Hebrew University, published in Nature Computational Science, introduces a new framework for interpreting single-cell and spatial omics data by monitoring deep neural networks training dynamics. The research aims to address the inherent ambiguities in cell annotations and offers a novel approach for understanding complex biological data.
Single-cell and spatial omics data have transformed our ability to explore cellular diversity and cellular behaviors in health and disease. However, the interpretation of these high-dimensional datasets is challenging, primarily due to the difficulty of assigning discrete and accurate annotations, such as cell types or states, to heterogeneous cell populations. These annotations are often subjective, noisy, and incomplete, making it difficult to extract meaningful insights from the data.
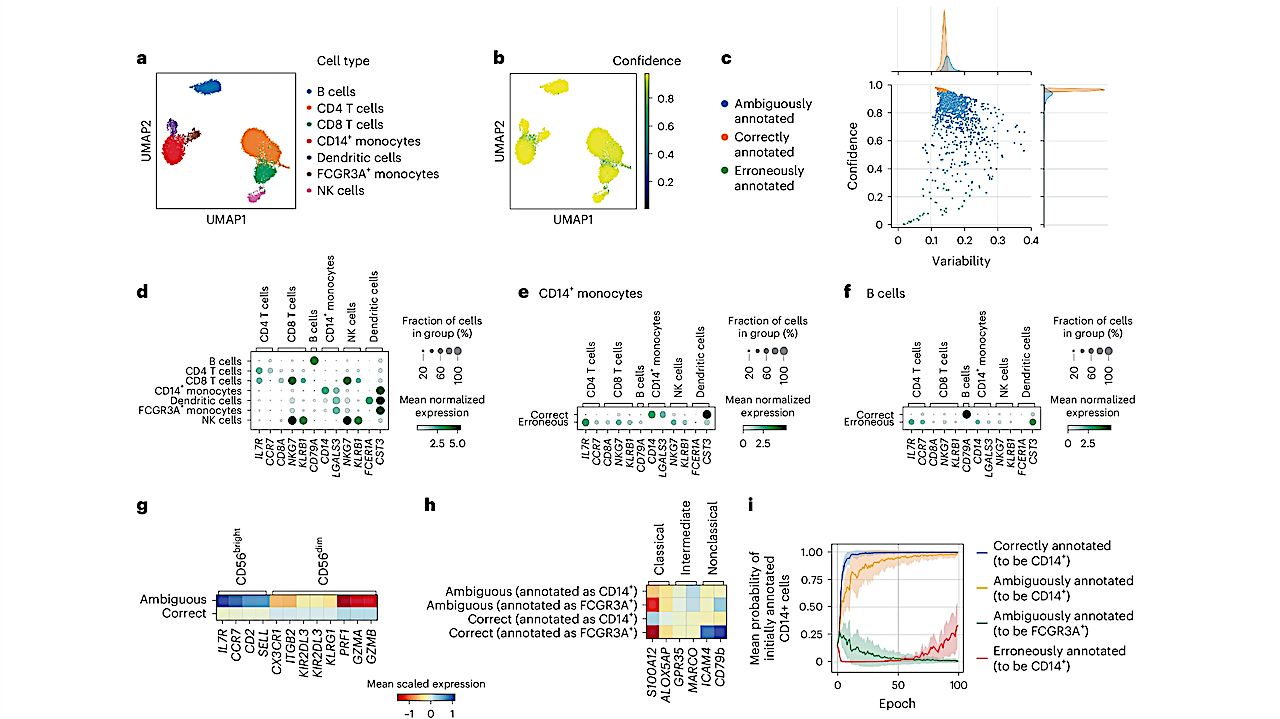
The researchers developed a new framework, Annotatability, which helps identify mismatches in cell annotations and better characterizes biological data structures. By monitoring the dynamics and difficulty of training a deep neural network over annotated data, Annotatability identifies areas where cell annotations are ambiguous or erroneous. The approach also highlights intermediate cell states and the complex, continuous nature of cellular development.
As part of the study, the team introduced a signal-aware graph embedding method that enables more precise downstream analysis of biological signals. This technique captures cellular communities associated with target signals and facilitates the exploration of cellular heterogeneity, developmental pathways, and disease trajectories.
The study demonstrates the applicability of Annotatability across a range of single-cell RNA sequencing and spatial omics datasets. Notable findings include the identification of erroneous annotations, delineation of developmental and disease-related cell states, and better characterization of cellular heterogeneity. The results highlight the potential of this framework for unraveling complex cellular behaviors and advancing our understanding of both health and disease at the single-cell level.
The researchers’ work presents a significant step forward in genomic data interpretation, offering a powerful tool for unraveling cellular diversity and enhancing our ability to study the dynamics of health and disease.
Interpreting single-cell and spatial omics data using deep neural network training dynamics, Nature Computational Science
Astrobiology, Genomics,