Structural Enzymology, Phylogenetics, Differentiation, and Symbolic Reflexivity at the Dawn of Biology
The reflexive translation of symbols in one chemical language to another defined genetics. Yet, the co-linearity of codons and amino acids is so commonplace an idea that few even ask how it arose.
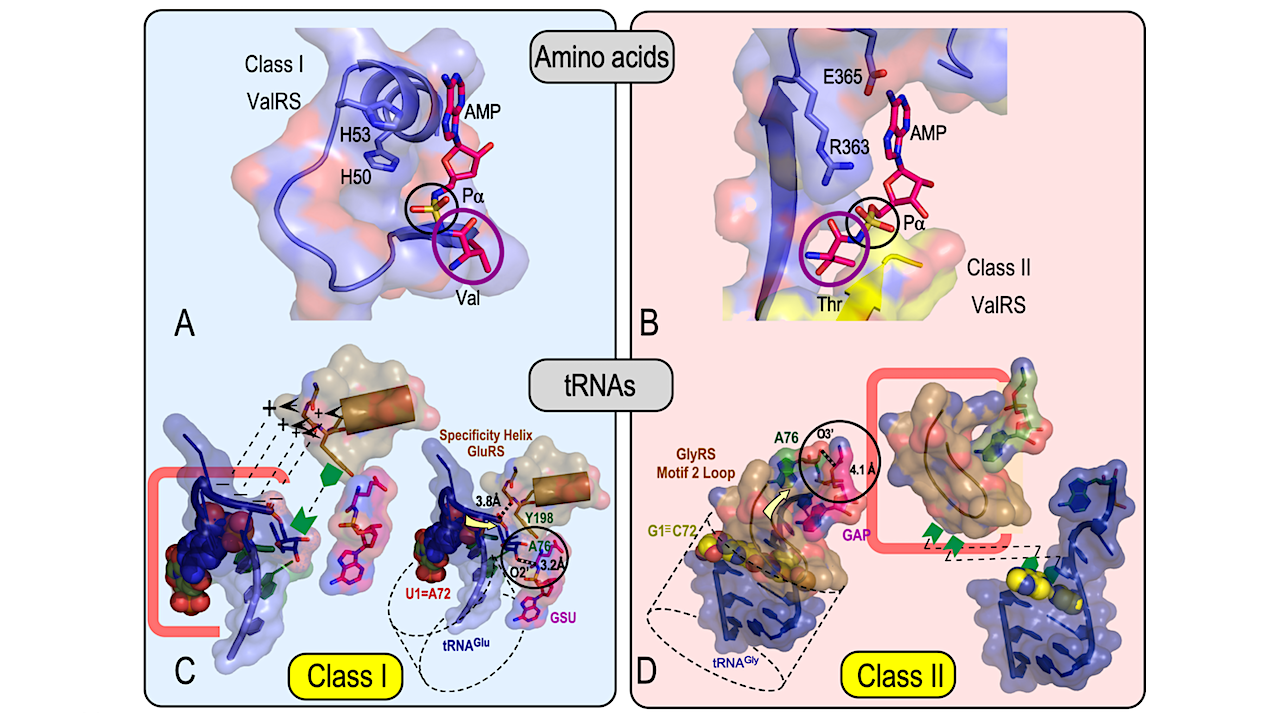
Readout is done by two distinct sets of proteins, called aminoacyl-tRNA synthetases (AARS). AARS must enforce the rules first used to assemble themselves. The roots of translation lie in experimentally testing the structural codes that the earliest AARS•tRNA cognate pairs used to recognize both amino acid and RNA substrates.
We review here new results on five different facets of that problem. (i) The surfaces of structures coded by opposite strands of the same gene have opposite polarities. The corresponding proteins then fold up “inside out” relative to one another. The inversion symmetry of base pairing thus projects into the proteome. That leads in turn to contrasting amino acid and RNA substrate binding modes.
(ii) E. coli reproduces in vivo the nested hierarchy of active excerpts we had designed as models—protozymes and urzymes—for ancestral AARS. (iii) A third novel deletion produced in vivo and a new Class II urzyme suggest how to design bidirectional urzyme genes.
(iv) Codon middle-base pairing provides a basis to constrain Class I and II AARS family trees. (v) AARS urzymes acylate Class-specific subsets of an RNA library, showing RNA substrate specificity for the first time. Four new phylogenetic routines augment these results to compose a viable platform for experimental study of the origins of genetic coding.
Significance Statement
The origin of genetic coding poses questions distinct from those faced studying the evolution of enzymes since the first cells. Modern enzymes that translate the code range in size from ∼330 to ∼970 amino acids.
Ancestral forms cannot have been nearly as complex. Moreover, such primitive enzymes likely could enforce only a much-reduced coding alphabet. Structural and molecular biology data point to a broad sketch of events leading to the code. That research platform will enable us to see how Nature came to store information about the physical chemistry of amino acids in the coding table.
That, in turn, allowed searching of a very broad amino acid sequence space. Selection could then learn how to assemble amino acids into functional, reflexive catalysts. Those catalysts had rates and fidelities consistent with bootstrapping the modern coding alphabet. New phylogenetic algorithms need to be developed to fully test that putative sketch experimentally.
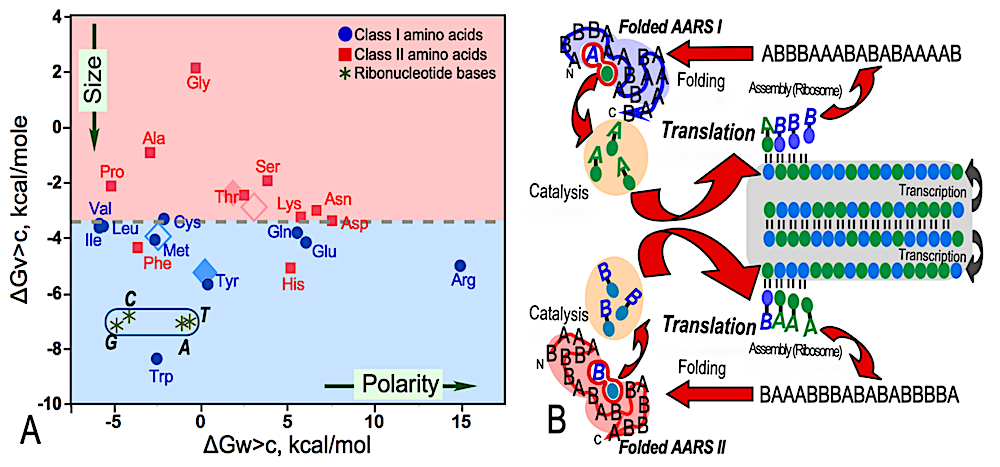
The origins of AARS reflexivity. A. Free energies of transfer for amino acids and ribonucleotide bases. These are the building blocks for proteins and nucleic acids. The Y axis is the free energy for transferring the side chain from vapor to cyclohexane. It is thus a surrogate for size. The X axis is the corresponding free energy for transferring the side chain from water to cyclohexane. It is a surrogate for polarity. The plot thus compares the physical chemistry of the nucleic acid and protein alphabets. Class I amino acids are blue dots; Class II amino acids are red squares. The colored background shows that Class I amino acids are predominantly bigger. They also span a larger range of polarity, although most are nonpolar Mean (solid) and median (outline) values for each Class are shown as diamonds of the same color. B. Schematic of the role of AARS in the information flow in genetics. A bidirectional ancestral gene and its mRNA transcripts are inside the gray panel. The gene is a bidirectional gene encoding Class I and II AARS on opposite strands. The respective translated peptides are written with a binary alphabet, with amino acids A, B activated respectively by Class I and Class II synthetases. Acylated RNAs are shown as capital letters linked to a green or blue ellipse, representing the symbolic codon representation. A folded conformation is essential for recognition of both amino acid and RNA substrates, and for stabilizing the two transition states for carboxyl group activation and acyl-transfer. Paired cycles of large red arrows define the reflexivity of AARS within each Class. Supplies of building blocks (acylated RNAs within amber ellipses) must be created by the two proteins, which must fold to catalyze the crucial reactions. Selection and gene replication are implicit in the cycle labeled “transcription”. — biorxiv.org
Structural Enzymology, Phylogenetics, Differentiation, and Symbolic Reflexivity at the Dawn of Biology, biorxiv.org (open access)
Astrobiology