Planetary Protection: Extremophilic Bacteria Isolated from the NASA Phoenix Mission Spacecraft Assembly Cleanrooms
Human-designed oligotrophic environments, such as cleanrooms, harbor unique microbial communities shaped by selective pressures like temperature, humidity, nutrient availability, cleaning reagents, and radiation.
During the Phoenix spacecraft mission, genomes of 215 bacterial isolates were sequenced and based on overall genome related indices, 53 strains belong to 26 novel species were recognized. Metagenome mapping indicated less than 0.1% of the reads associated with novel species, suggesting their rarity.
Genes responsible for biofilm formation, such as BolA (COG0271) and CvpA (COG1286), were predominantly found in proteobacterial members but were absent in other non-spore-forming and spore-forming species. YqgA (COG1811) was detected in most spore-forming members but was absent in Paenibacillus and non-spore-forming species. Cell fate regulators, COG1774 (YaaT), COG3679 (YlbF, YheA/YmcA), and COG4550 (YmcA, YheA/YmcA), controlling sporulation, competence, and biofilm development processes, were observed in all spore-formers but were missing in non-spore-forming species.
COG analyses further revealed resistance-conferring proteins in all spore-forming novel species (n=13) and eight actinobacteria, responsible for enhanced membrane transport and signaling under radiation (COG3253), transcription regulation under radiation stress (COG1108), and DNA repair and stress responses (COG2318).
Additional functional analysis revealed that Agrococcus phoenicis, Microbacterium canaveralium, and Microbacterium jpeli contained biosynthetic gene clusters (BGCs) for ε-Poly-L-lysine, beneficial in food preservation and biomedical applications. Two novel Sphingomonas species exhibited zeaxanthin, an antioxidant beneficial for eye health.
Paenibacillus canaveralius harbored genes for bacillibactin, crucial for iron acquisition. Georgenia phoenicis had BGCs for alkylresorcinols, compounds with antimicrobial and anticancer properties used in food preservation and pharmaceuticals. Despite strict decontamination and controlled environmental conditions, cleanrooms foster novel bacterial species which can form biofilms, resist various stressors, and produce biotechnologically valuable compounds.

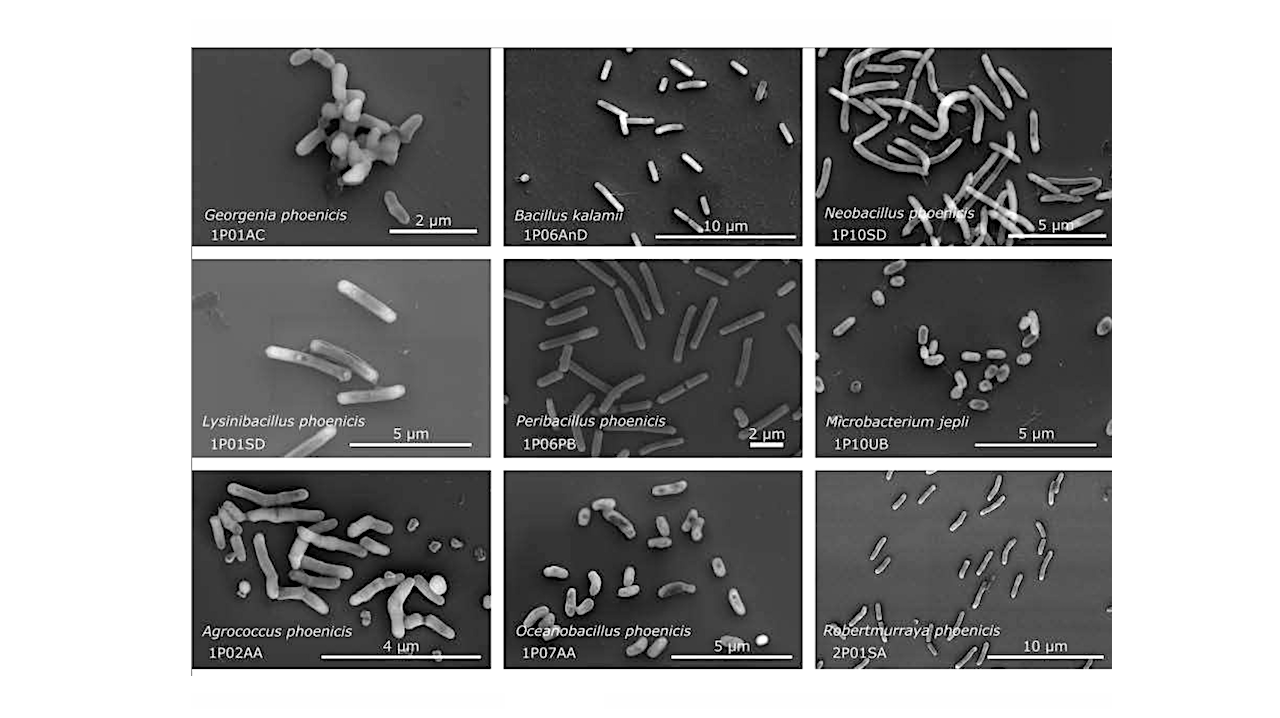
Scanning electron microscopy of the novel species isolated from the Phoenix spacecraft 730 assembly cleanroom. — Research Square
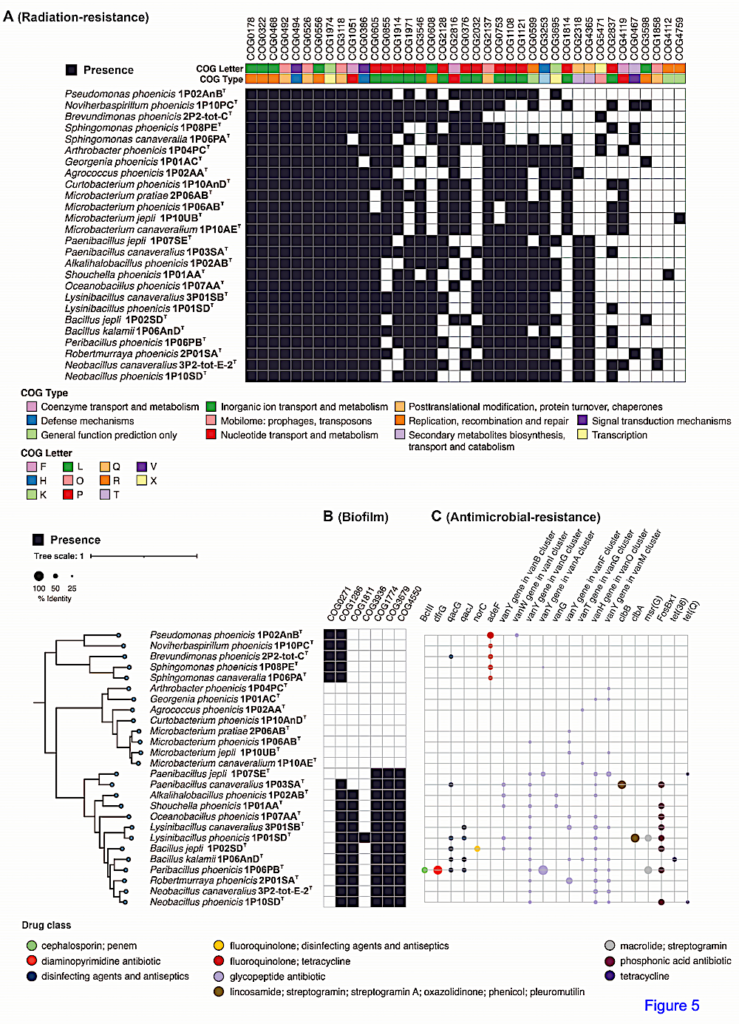
Functional insights into novel species from NASA cleanrooms. A. Presence of radiation resistance COGs (from Pal et al.87) in the 26 novel species, revealing their genetic potential for radiation resilience. B. Identification of Proteins of Interest (PoI) using KMAP with applications in various industries, leveraging the unique functional capabilities of novel species from NASA cleanrooms. — Research Square

Comparative analysis of ε-Poly-L-lysine synthetase in novel species. A. ε-Poly-L-lysine gene cluster comparison in Epichloe festucae (fungal producer), Corynebacterium variabile (bacterial producer), and three novel species from our study (Agrococcus phoenicis, Microbacterium canaveralium, Microbacterium jepli) and Leifsonia virtsii (isolated from ISS) show conserved gene cluster architecture. B. Protein sequence alignment of ε-Poly-L-lysine synthetase enzymes from these organisms exhibits conserved domains, including NRPS adenylation (A), thiolation (T), transmembrane (TM), and C- terminal tandem domains (C1, C2, C3). — Research Square
Júnia Schultz, King Abdullah University of Science and Technology; Tahira Jamil, King Abdullah University of Science and Technology; Pratyay Sengupta, Indian Institute of Technology Madras; Shobhan Karthick Muthamilselvi Sivabalan, Indian Institute of Technology Madras; Anamika Rawat, King Abdullah University of Science and Technology; Niketan Patel, King Abdullah University of Science and Technology; Srinivasan Krishnamurthy,Institute of Microbial Technology; Intikhab Alam, King Abdullah University of Science and Technology; Nitin Kumar Singh, Jet Propulsion Laboratory; Karthik Raman, Indian Institute of Technology Madras; Alexandre Soares Rosado, King Abdullah University of Science and Technology; Kasthuri Venkateswaran, Jet Propulsion Laboratory
Functional Insights into Novel Extremophilic Bacteria Isolated from the NASA Phoenix Mission Spacecraft Assembly Cleanrooms, Research Square (open access)
This work is licensed under a CC BY 4.0 License
Astrobiology