Urea Assimilation and Oxidation Supports the Activity of a Phylogenetically Diverse Microbial Community in the Dark Ocean
Urea is hypothesized to be an important source of nitrogen and chemical energy to microorganisms in the deep sea; however, direct evidence for urea use below the epipelagic ocean is lacking.
Here, we explore urea utilization from 50 to 4000 meters depth in the northeastern Pacific Ocean using metagenomics, nitrification rates, and single-cell stable-isotope-uptake measurements with nanoscale secondary ion mass spectrometry (nanoSIMS). We find that the majority (>60%) of active cells across all samples assimilated urea-derived N, and that cell-specific nitrogen-incorporation rates from urea were higher than that from ammonium.
Both urea concentrations and assimilation rates relative to ammonium generally increased below the euphotic zone. We detected ammonia- and urea-based nitrification at all depths at one of two sites analyzed, demonstrating their potential to support chemoautotrophy in the mesopelagic and bathypelagic regions.
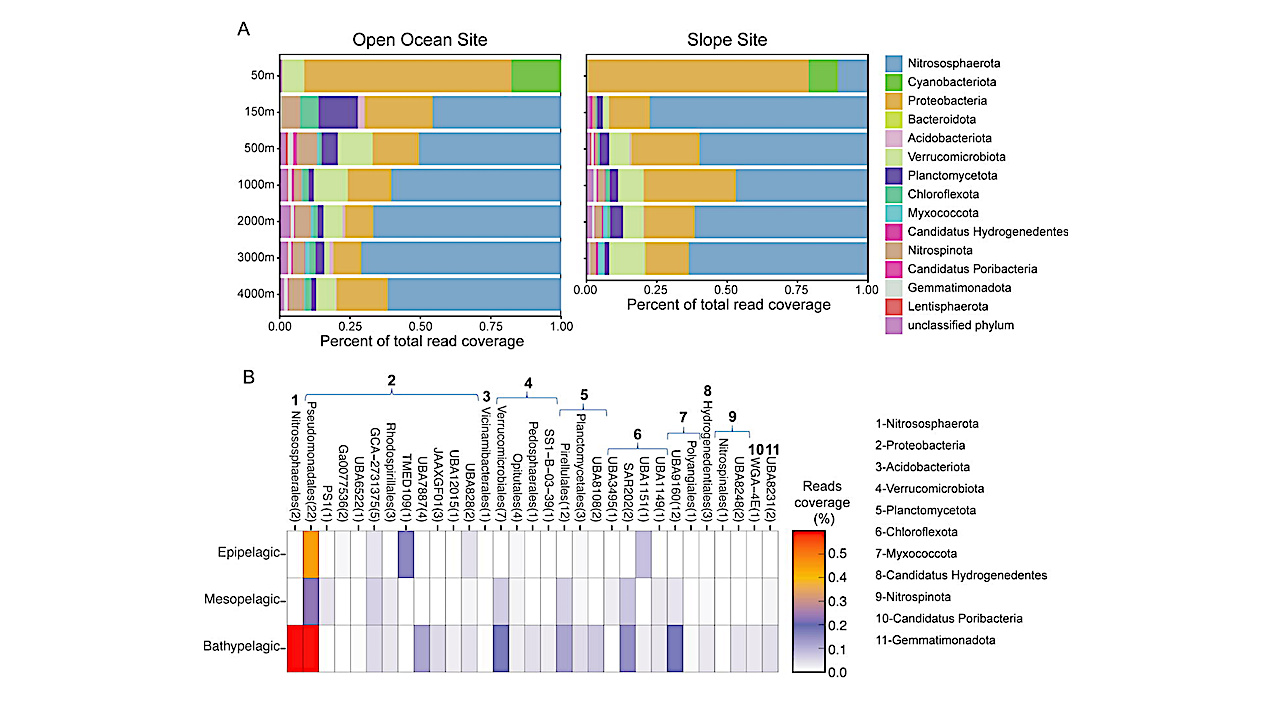
Using newly generated metagenomes we find that the ureC gene, encoding the catalytic subunit of urease, is found within 39% of deep-sea cells in this region, including the Nitrosophaerota (likely for nitrification) as well as thirteen other phyla such as Proteobacteria, Verrucomicrobia, Plantomycetota, Nitrospinota, and Chloroflexota (likely for assimilation).
Analysis of public metagenomes revealed ureC within 10-46% of deep-sea cells around the world, with higher prevalance below the photic zone, suggesting urea is widely available to the deep-sea microbiome globally. Our results demonstrate that urea is a nitrogen source to abundant and diverse microorganisms in the dark ocean, as well as a significant contributor to deep-sea nitrification and therefore fuel for chemoautotrophy.
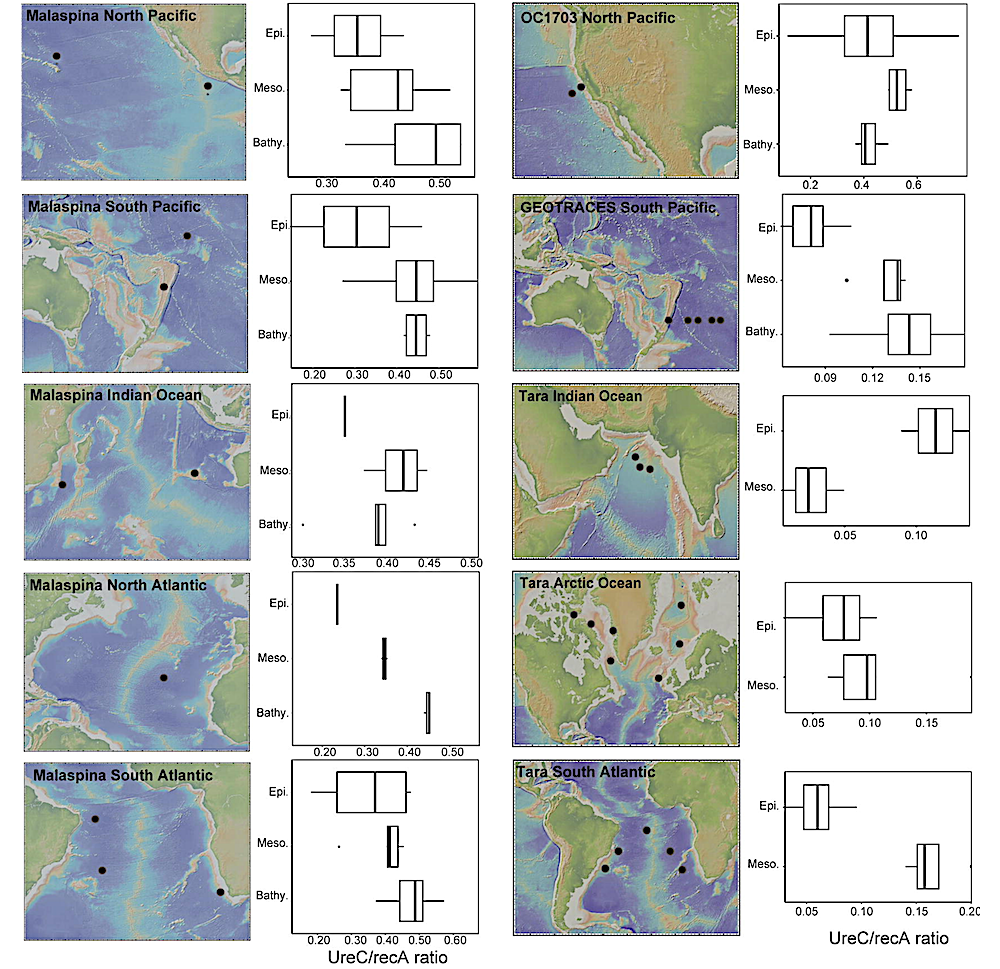
Box plots showing abundance of ureC genes relative to total mapped reads in the 701 epipelagic (0-200 mbsl), mesopelagic (200-1000 mbsl), and bathypelagic (1000-4500 mbsl) available under aCC-BY-NC-ND 4.0 International license. was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made bioRxiv preprint doi: https://doi.org/10.1101/2024.07.26.605319; this version posted July 27, 2024. The copyright holder for this preprint (which regions of different ocean regions. The North Pacific Ocean is shown in the OC1703 and Malaspina datasets, Southwest Pacific Ocean in the GEOTRACES dataset, North Atlantic in the Malaspina dataset, South Atlantic and Indian Oceans in the Malaspina and Tara Oceans datasets and the Arctic Ocean in the Tara Oceans datasets. Gene abundances are displayed as the ratio between ureC and recA (both gene coverages calculated as RPKM). For the Tara Oceans, only samples for the epipelagic and mesopelagic regions were available. — biorxiv.org
Nestor Arandia-Gorostidi, Alexander L. Jaffe, Alma E. Parada, Bennett J. Kapili, Karen L. Casciotti, Rebecca S. R. Salcedo, Chloé M. J. Baumas, Anne E. Dekas
Astrobiology,