Structure-based Inference Of Eukaryotic Complexity In Asgard Archaea
Asgard archaea played a key role in the origin of the eukaryotic cell. While previous studies found that Asgard genomes encode diverse eukaryotic signature proteins (ESPs), representing homologs of proteins that play important roles in the complex organization of eukaryotic cells, the cellular characteristics and complexity of the Asgard archaeal ancestor of eukaryotes remain unclear.
Here, we used de novo protein structure modeling and sensitive sequence similarity detection algorithms within an expanded Asgard archaeal genomic dataset to build a structural catalogue of the Asgard archaeal pangenome and identify 908 new isomorphic ESPs (iESPs), representing clusters of protein structures most similar to eukaryotic proteins and that likely underwent extensive sequence divergence.
While most previously identified ESPs were involved in cellular processes and signaling, iESPs are enriched in information storage and processing functions, with several being potentially implicated in facilitating cellular complexity.
By expanding the complement of eukaryotic proteins in Asgard archaea, this study indicates that the archaeal ancestor of eukaryotes was more complex than previously assumed.

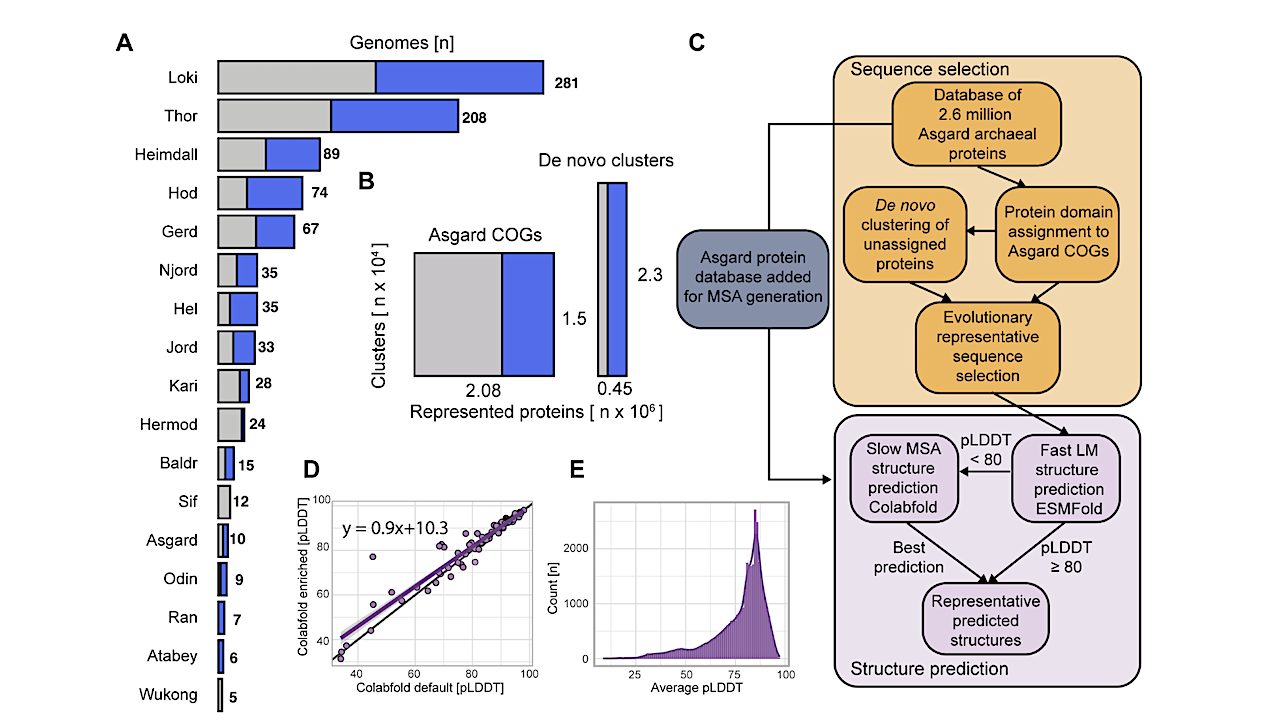
Structure-guided identification of functionally diverse iESP structural clusters. (A) Workflow to cluster protein structures and identify iESPs. (B) Identification of Asgard archaeal iESPs based on structural similarity. (C) Bar chart summarizing the clustering of previously described ESP and iESP protein structures into structural clusters, respectively. (D) Sankey diagram displaying functional categories of newly identified iESPs clusters and clusters containing previously established ESPs. Categories are inferred from the best SwissProt hits EggNOG annotation. ‘Multiple’ indicates an association of a structural cluster with multiple functional categories. (E) Subgraph of protein structure similarity network, highlighting small GTPase (black outline) and Argonaute proteins. P, prob ability. — biorxiv.org
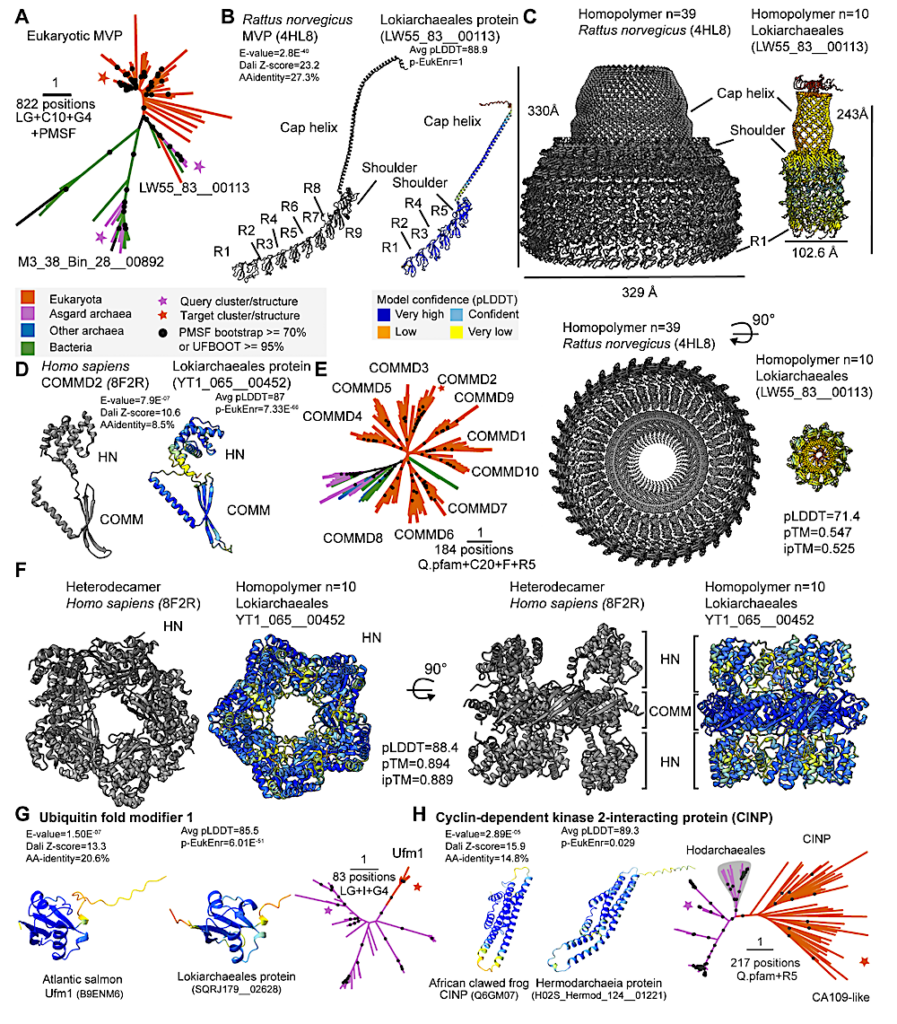
Asgard archaeal protein complexes implicating cellular compartmentalization. Asgard archaeal proteins related to eukaryotic (A-C) MVPs and (D-F) COMMD-containing proteins. (A) Phylogeny of prokaryotic and eukaryotic full-length MVPs. See Fig. S4A for tree based only on the shoulder domain. (B) Rat MVP complex (45) next to Lokiarchaeial MVP (predicted structure) indicating the cap helix, shoulder, and repeat domains (R). (C) Biological assembly of the rat MVP cap (left) next to5 a multimer model of the Asgard archaeal homodecamer (right). (D) Human COMMD2 next to Lokiarchaeial homolog indicating the HN and COMM domains. (E) Phylogeny of prokaryotic and eukaryotic COMMD-containing proteins. (F) Resolved human COMMD heterodecamer (46) next to a multimer model of the Asgard archaeal homodecamer. (G, H) Identification of Asgard archaeal iESPs of eukaryotic ubiquitin fold modifier 1 (G) and cyclin-dependent kinase 2-interacting protein (Hodarchaeales10 clade indicated with grey background) (H). Asgard archaeal query protein structure, best-scoring SwissProt target structural model and phylogenetic analysis of related protein sequences are indicated in the left, middle and right panel, respectively. Structural models exclude long terminal disordered regions. Additional data include Foldseek E-value, Dali Z-score, enrichment of eukaryotic structures (Fisher’s exact test, Bonferroni-corrected p-value, ‘p-EukEnr’), and amino-acid identity to best structure hit (‘AA-15 identity’). Phylogenetic analyses highlight sequences for query and target structures, input MSA positions, and substitution model. Scale bar: 1 amino acid substitution per position. Multimer model confidence measures (pLDDT, pTM, ipTM) are indicated. — biorxiv.org
Structure-based inference of eukaryotic complexity in Asgard archaea, biorxiv.org
Astrobiology