Oxygen Metabolism In Descendants Of The Archaeal-Eukaryotic Ancestor
Asgard archaea were pivotal in the origin of complex cellular life. Hodarchaeales (Asgardarchaeota class Heimdallarchaeia) were recently shown to be the closest relatives of eukaryotes. However, limited sampling of these archaea constrains our understanding of their ecology and evolution, including their anticipated role in eukaryogenesis.
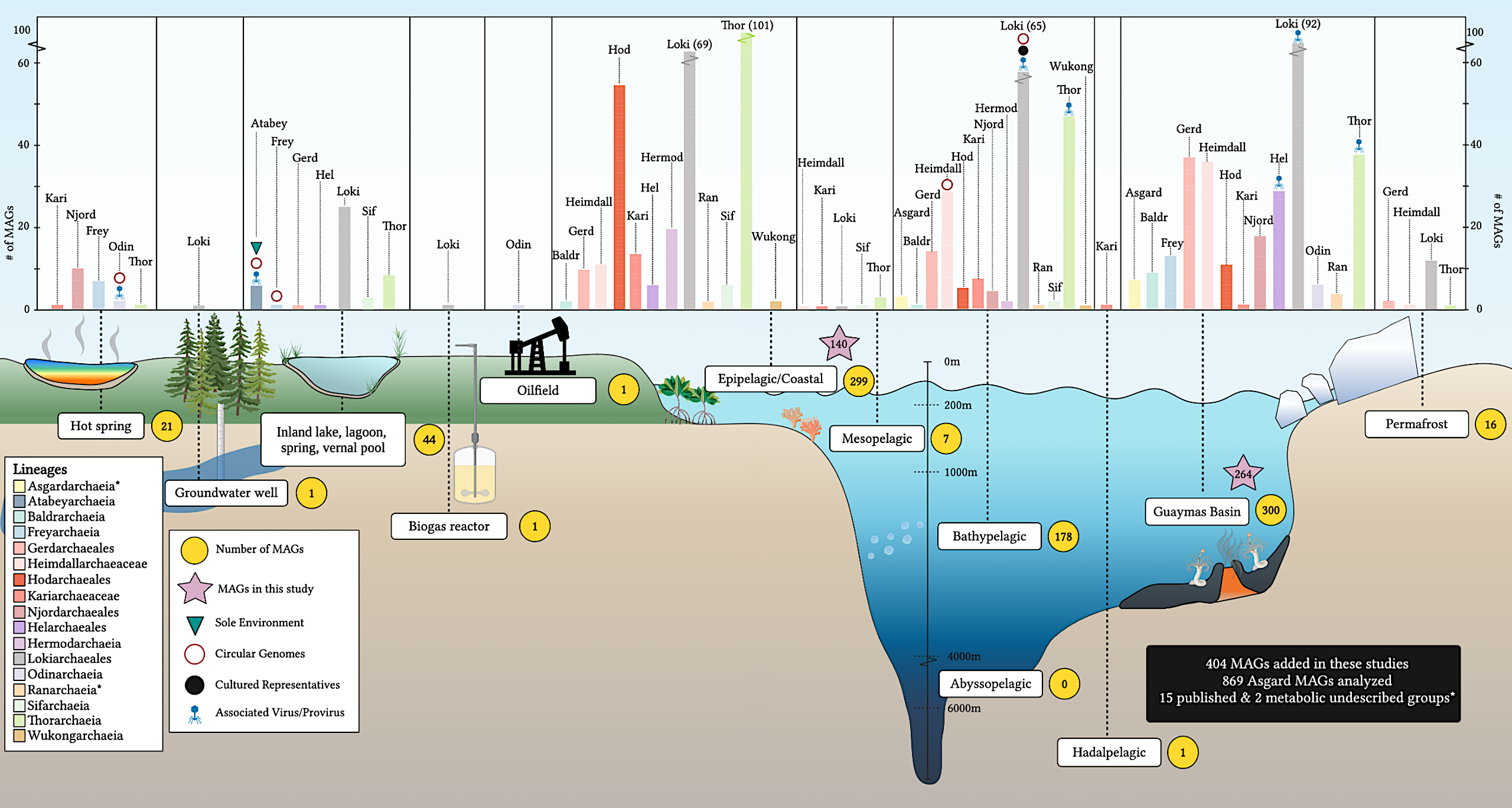
Here, we nearly double the number of Asgardarchaeota metagenome-assembled genomes (MAGs) to 869, including 136 new Heimdallarchaeia (49 Hodarchaeales) and several novel lineages. Examining global distribution revealed Hodarcheales are primarily found in coastal marine sediments. Detailed analysis of their metabolic capabilities revealed guilds of Heimdallarchaeia are distinct from other Asgardarchaeota. These archaea encode hallmarks of aerobic eukaryotes, including electron transport chain complexes (III and IV), biosynthesis of heme, and response to reactive oxygen species (ROS).
The predicted structural architecture of Heimdallarchaeia membrane-bound hydrogenases includes additional Complex-I-like subunits potentially increasing the proton motive force and ATP synthesis. Heimdallarchaeia genomes encode CoxD, which regulates the electron transport chain (ETC) in eukaryotes. Thus, key hallmarks for aerobic respiration may have been present in the Asgard-eukaryotic ancestor.
Moreover, we found that Heimdallarchaeia is present in a variety of oxic marine environments. This expanded diversity reveals these Archaea likely conferred energetic advantages during early stages of eukaryogenesis, fueling cellular complexity.
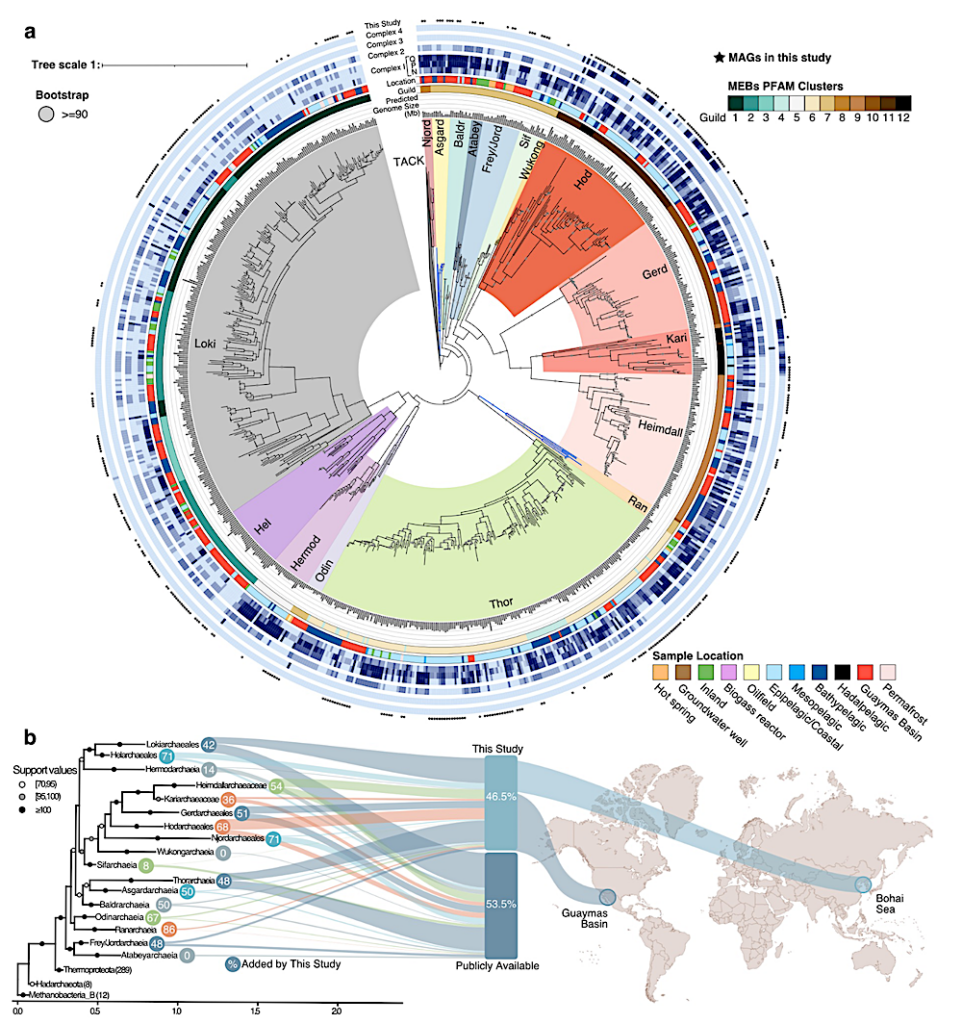
Expanded diversity of Asgard archaea. a. Maximum likelihood phylogeny based on 47 arCOGs for the 837 Asgard MAGs and 36 TACK archaea representatives as the outgroup. The tree was generated using RAxML v8.2.11 with free model parameters estimated by the RAxML GAMMA model of rate heterogeneity and ML estimate of alpha-parameter–bootstraps based on 1000 rapid bootstrap inferences. The blue branches (lower right) indicate the new Asgararchaeota class, Ranarchaeia and the recently proposed Asgardarchaeia (top). The concentric rings (in-out) highlight the predicted genome size (Mb), metabolic guilds based on Pfam clustering, sampling locations, subunits in the electron transport chain Complex I-IV (Complex I modules are shown separately), and black stars for the MAGs added by this study. b. Mapped on the phylogeny, using the new NM47 markers (see methods), is the percentage of MAGs from each lineage added by this study (circle), including 46.5% of the catalog from the Guaymas Basin (65.3%) and the Bohai Sea (34.7%). These MAGs constitute a 115.1% increase in the medium-high quality publicly available genomes — biorxiv.org

Updated Heimdallarchaiea-centric model of eukaryogenesis in light of an expanded catalog of Asgard genomes. Our data suggest the Last Asgard archaea Common Ancestor (LAsCA) diversification allowed widespread niche expansion, including deep-sea associated lineages, Ranarchaeia (Ran; orange) and Asgardarchaeia (Asgard; yellow). Nuo subunits in several Asgardarchaeota indicate an earlier transition from MBH to Complex-I-like respiratory systems. Exposure to oxygen and the byproducts of aerobic respiration before the Great Oxidation Event triggered the exchange of oxygen tolerance and increasing interactions with the aerobic mitochondrial ancestor (blue). In Heimdallarchaeia (Heimall; red), we identify the loss of nitrate reduction (NarGHJ) and vitamin B12 (vB12) synthesis in coastal genomes present in the deep sea MAGs. The gain of ETC complexes (III and IV) increased energy yield and reactive oxygen species (ROS) production. Metabolite exchanges and coordinated response to rising oxygen levels may have increased cell-to-cell contact between partners, enabling initial compartmentalization. The Heimdallarchaeia host increased cellular regulation through ESPs (ORML, JAKMIP, L28e), using protoglobin to bind oxygen, the terminal electron acceptor. Asgard host-encoded CoxD could have controlled the endosymbiont’s ROS production in the first eukaryotic common ancestor (FECA). By the last eukaryotic common ancestor (LECA), endosymbiont gene transfer (EGT) relocated mitochondrial-encoded ETC subunits to the nuclear genome (nDNA), catalyzing the diversity of modern eukaryotes. The red arrow highlights potential regulation of ETC byproducts (ROS and ATP) by CoxD in the host’s genome. Today’s Hodarchaeales and Kariarchaeaceae are aerobic chemoheterotrophs, co-occurring with Alphaproteobacteria in oxic coastal sediments (see the main text for details). Additional abbreviations include OM (organic matter), NO red (nitrate reduction), ESP (eukaryotic signature protein), EMP (Embden–Meyerhof–Parnas Pathway), ED (Entner-Doudoroff Pathway), O-PPP (Oxidative Pentose Phosphate Pathway, NO-PPP (Non-oxidative Pentose Phosphate Pathway), pTCA (partial Tricarboxylic Acid) Cycle, and WLP (Wood-Ljungdahl Pathway). — biorxiv.org
Oxygen metabolism in descendants of the archaeal-eukaryotic ancestor, biorxiv.org
Astrobiology