Offworld Life Science Hardware: Real-Time Culture-Independent Microbial Profiling Onboard the International Space Station Using Nanopore Sequencing
For the past two decades, microbial monitoring of the International Space Station (ISS) has relied on culture-dependent methods that require return to Earth for analysis.
This has a number of limitations, with the most significant being bias towards the detection of culturable organisms and the inherent delay between sample collection and ground-based analysis. In recent years, portable and easy-to-use molecular-based tools, such as Oxford Nanopore Technologies’ MinION™ sequencer and miniPCR bio’s miniPCR™ thermal cycler, have been validated onboard the ISS.
Here, we report on the development, validation, and implementation of a swab-to-sequencer method that provides a culture-independent solution to real-time microbial profiling onboard the ISS. Method development focused on analysis of swabs collected in a low-biomass environment with limited facility resources and stringent controls on allowed processes and reagents.

Workflow of the in-situ culture-independent, swab-to-sequencer process for extreme environment microbial profiling. (A) Swabbing of a 100 cm2 area. (B) Insertion of the swab head directly into lysis buffer in a PCR tube and placement into miniPCR for DNA extraction. (C) Bead-based DNA purification (D) DNA amplification within miniPCR. (E) Bead-based PCR amplicon purification. (F) Addition of sequencing control DNA and ONT-based rapid adaptor library preparation. (G) Flow cell loading and MinION sequencing — Genes .
ISS-optimized procedures included enzymatic DNA extraction from a swab tip, bead-based purifications, altered buffers, and the use of miniPCR and the MinION. Validation was conducted through extensive ground-based assessments comparing current standard culture-dependent and newly developed culture-independent methods.
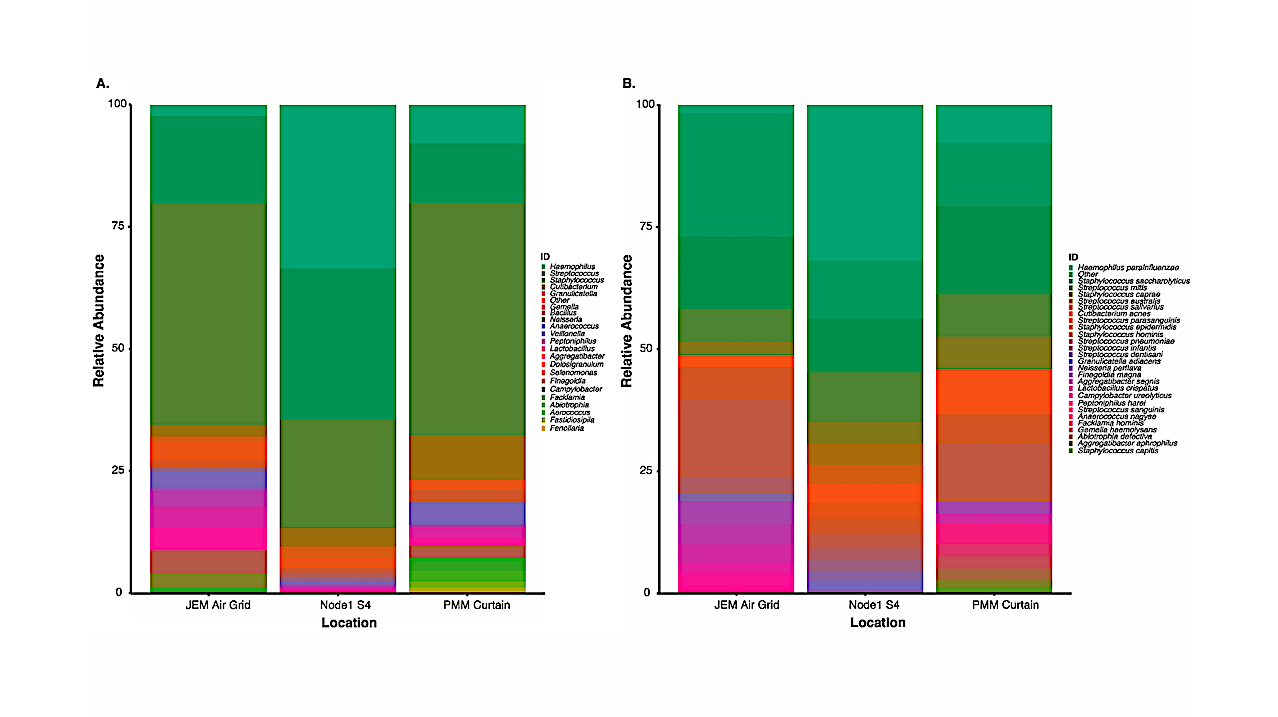
Similar microbial distributions were observed between the two methods; however, as expected, the culture-independent data revealed microbial profiles with greater diversity. Protocol optimization and verification was established during NASA Extreme Environment Mission Operations (NEEMO) analog missions 21 and 22, respectively.
Unique microbial profiles obtained from analog testing validated the swab-to-sequencer method in an extreme environment. Finally, four independent swab-to-sequencer experiments were conducted onboard the ISS by two crewmembers. Microorganisms identified from ISS swabs were consistent with historical culture-based data, and primarily consisted of commonly observed human-associated microbes.
This simplified method has been streamlined for high ease-of-use for a non-trained crew to complete in an extreme environment, thereby enabling environmental and human health diagnostics in real-time as future missions take us beyond low-Earth orbit.
Real-Time Culture-Independent Microbial Profiling Onboard the International Space Station Using Nanopore Sequencing, Genes (open access)

Location of swabbing onboard the International Space Station. (A). On July 19, 2018 the Node 1S4 wall adjacent to the crew dining table was swabbed. The area swabbed is directly behind the spoon seen attached to the Node 1S4 interior wall. (B). On September 17, 2018 a swab sample was collected from a curtain within the Permanent Multipurpose Module (PMM) that separates the PMM module from the Node 3 module. The area swabbed faces into the PMM. (C). On September 24, 2018 a swab sample was collected from an area of the Overhead Aft 7 Air Grid in the Japanese Experiment Module (JEM) — Genes .
Astrobiology