Prebiotic Gas Flow Environment Enables Isothermal Nucleic Acid Replication

Nucleic acid replication is a central process at the origin of life. On early Earth, replication is challenged by the dilution of molecular building blocks and the difficulty of separating daughter from parent strands, a necessity for exponential replication.
While thermal gradient systems have been shown to address these problems, elevated temperatures lead to degradation. Also, compared to constant temperature environments, such systems are rare. The isothermal system studied here models an abundant geological environment of the prebiotic Earth, in which water is continuously evaporated at the point of contact with the gas flows, inducing up-concentration and circular flow patterns at the gas-water interface through momentum transfer.
We show experimentally that this setting drives a 30-fold accumulation of nucleic acids and their periodic separation by a 3-fold reduction in salt and product concentration. Fluid dynamic simulations agree with observations from tracking fluorescent beads. In this isothermal system, we were able to drive exponential DNA replication with Taq polymerase. The results provide a model for a ubiquitous non-equilibrium system to host early Darwinian molecular evolution at constant temperature.
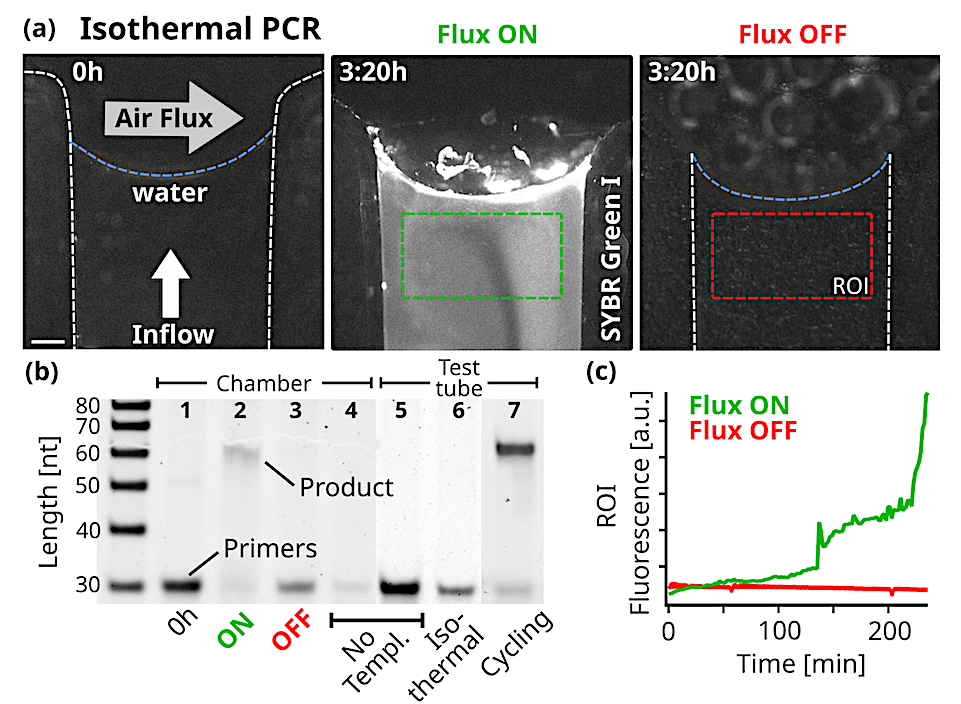
Replication. (a) Fluorescence micrographs of the PCR reaction in the chamber. At isothermal 68°C, 10 µl of reaction sample was subjected to a constant 5 nl/s pure water flow towards the interface where a 250ml/min gas flowed perpendicularly. The initial state on the left shows the background fluorescence. Fluorescence increased under flux (middle, after 3:20h), while without flux the fluorescence signal remained minimal (right). The reaction sample consisted of 0.25 µM primers, 5 nM template, 200 µM dNTPs, 0.5 X PCR buffer, 2.5 U Taq polymerase, 2 X SYBR Green I. Scale bar is 250 µm. (b) 15% Polyacrylamide Gel Electrophoresis of the reactions and neg. controls. After 4 hours in the reaction chamber with air- and waterflux ON, the 61mer product was formed under primer consumption (2), unlike in the equivalent experiment with the fluxes turned OFF (3). At the beginning of the experiment (1) or in the absence of template (4), no replicated DNA was detected. The reaction mixture was tested by thermal cycling in a test tube (5-7). As expected, replicated DNA was detected only with the addition of template: (7) shows the sample after 11 replication cycles. The sample was also incubated for 4 hours at the chamber temperature (68°C) yielding no product (6). Primer band intensity variations are caused by material loss during extraction from the microfluidic chamber. (c) SYBR Green I fluorescence increased when gas and water flow were turned on, but remained at background levels without flow. Fluorescence was averaged over time from the green and red regions of interest shown in (a). SYBR Green I fluorescence indicates replication, as formed products are able to hybridize. — biorxiv.org
Prebiotic gas flow environment enables isothermal nucleic acid replication, biorxiv.org
Astrobiology