NASA GeneLab New Workflows for Processing Microarray Data

Microarray technology enables simultaneous analysis of thousands of genes, facilitating understanding of complex diseases and molecular responses to environmental factors such as microgravity in space biology experiments.
Recently, GeneLab has wrapped the Microarray Affymetrix processing pipeline and the Microarray Agilent 1-channel processing pipeline into Nextflow workflows.
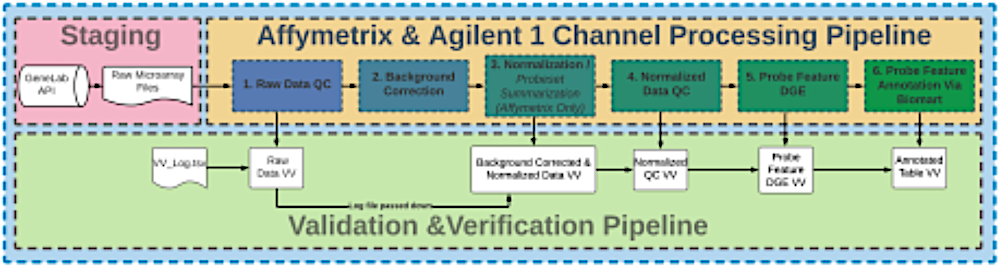
Nextflow is a workflow manager that enables reproducible and scalable workflows using software containers such as Docker and Singularity-a standalone, executable package that includes everything needed to run a piece of software, such as code, runtime, system tools, libraries, and settings. The workflows each include three subworkflows:
- The Analysis Staging subworkflow, which extracts metadata and raw files from the Open Science Data Repository (OSDR).
- The Microarray Affymetrix or Agilent 1-channel Pipeline subworkflow, which processes the data, accordingly.
- The V&V Pipeline subworkflow, which performs real-time validation and verification (V&V) on the processed data files.
The Affymetrix and Agilent 1-channel workflows are available for download, along with installation instructions, on the GeneLab Data Processing (DP) GitHub Repository and are instrumental for the GeneLab data processing team to process Microarray data hosted on the OSDR. So, if you are working with microarray data, then check out these latest workflows!
More info https://genelab.nasa.gov/workflows_microarray
Astrobiology, space biology, genomics,