Genomics Tricorder Tech: De novo Genome Sequencing And Annotation Of A Thermophilic Cyanobacterium Using The Oxford Nanopore Technologies Sequencing Platform

Editor’s note: Oxford Nanopore technology has been used multiple times on the International Space Station (see Genomics, Proteomics, Bioinformatics) to try and help elucidate changes in genomic function during exposure to conditions of spaceflight. This technology has also been used to track down the source of diseases in remote areas, characterize extremophiles in Antarctica, and study sources of food contamination. As we prepare for Away Team traverses on other worlds advanced versions of this sort of sequencing technology will be essential components of a crew’s toolset.
In this project the MinION DNA sequencing platform was tested and introduced. It was proven that Next-Generation Sequencing (NGS) is not restricted to core facilities, but can be implemented in a standard laboratory setting.
This makes the MinION suitable for decentralized strain screening, development and quality monitoring applications in the biotechnology industry. Over the course of the MinION early Access Program, the device was used to sequence a novel extremophile cyanobacteria candidate isolated from a hot spring at Klein Barmen, Namibia. The de novo assembly of the microorganism genome resulted in one bacterial chromosome without gaps. Further analysis showed, that significant variation/heterogeneity between related genomes is present. This underlines the novel character of the sequenced genome.
Recent advances in next-generation sequencing technology, have enabled decentralized high27 throughput DNA sequencing. Oxford Nanopore Technologies™ is the first to have made a long-awaited nanopore DNA sequencer available to a broader scientific audience. The company selected a number of researchers (~1000) for an initial beta test and developmental phase. Which was the initial foundation for this work. Being a member of the MinION™ device early access community (MAP) was a great honour. This chapter will focus on the fundamental theoretical background in order to make the presented work comprehensible. The first section starts with DNA sequencing history, thereafter moving on to the Oxford Nanopore™ MinION™ the DNA Sequencing device. The minute sequencing platform and its performance will be presented and discussed throughout the study. At the end of this chapter the cyanobacteria phylum will be introduced.
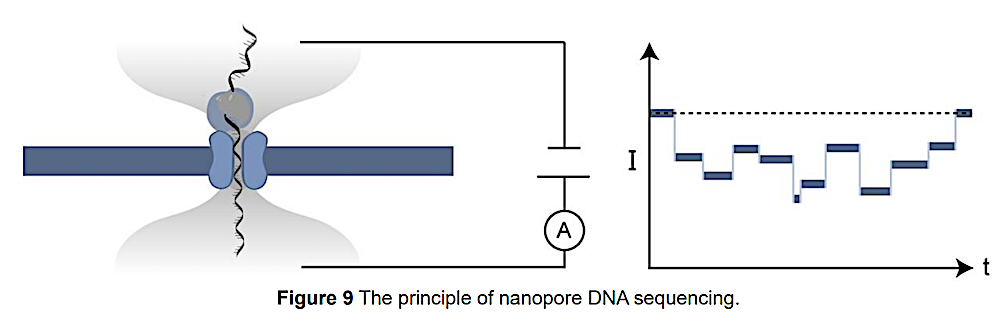
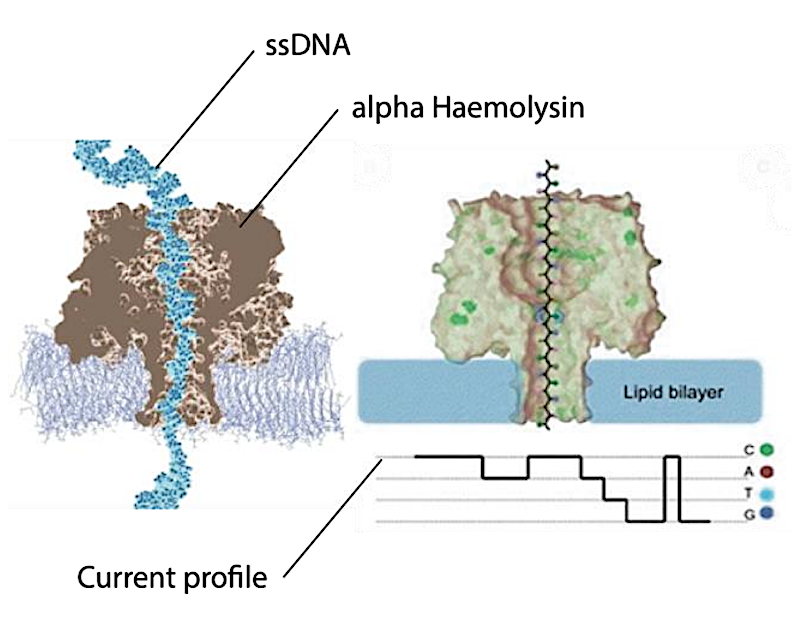
MD simulation of the 232 kDa Alpha- Haemolysin with ssDNA from Klaus Schulten et al. and to the right an illustration of the pore and the resulting current profile from Wang.

The Oxford Nanopore Technologies™ MinION™ device. Left: Workstation with handheld DNA sequencing platform. Right top: Components of the ONT Sequencing Kit for the library prep. Right bottom: Lab journal with a paper based checklist for optimal documentation and error reduced Library preparation. A tool which was developed together with ONT during the study.
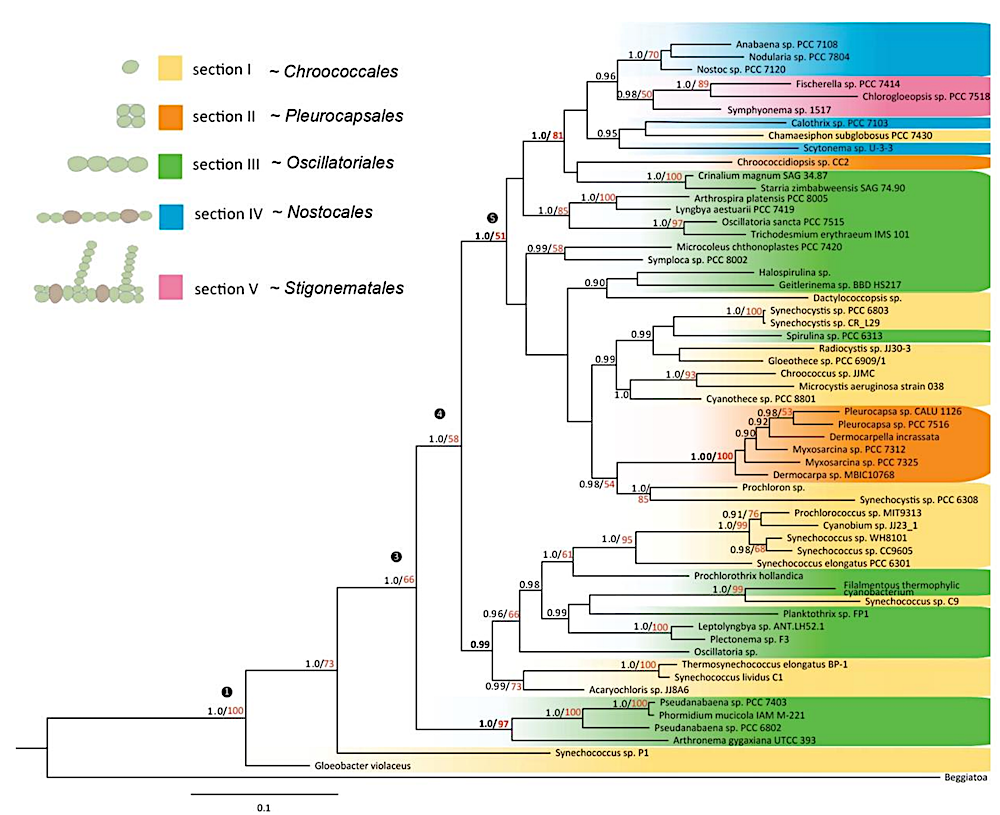
Phylogenetic tree of sequenced cyanobacteria (modified with Adobe AI).
Astrobiology