Tricorder Tech: Air Monitoring By Nanopore Sequencing
Editor’s note: Upon entering a spacecraft or arriving on a new, inhabited world, you would probably like to have your tricorder to have the ability to do a quick assay of the atmosphere that you and your team may soon be breathing. Here is one approach to consider as we design these tricorders that would use nanopore shotgun sequencing.
While the air microbiome and its diversity are known to be essential for human health and ecosystem resilience, the current lack of holistic air microbial diversity monitoring means that little is known about the air microbiome’s composition, distribution, or functional effects.
We here show that nanopore shotgun sequencing can robustly assess the air microbiome in combination with active air sampling through liquid impingement and tailored computational analysis. We provide laboratory and computational protocols for air microbiome profiling, and reliably assess the taxonomic composition of the core air microbiome in controlled and natural environments.
Based on de novo assemblies from the long sequencing reads, we further identify the taxa of several air microbiota down to the species level and annotate their ecosystem functions, including their potential role in natural biodegradation processes.
As air monitoring by nanopore sequencing can be employed in an automatable, fast, and portable manner for in situ applications all around the world, we envision that this approach can help holistically explore the role of the air microbiome.
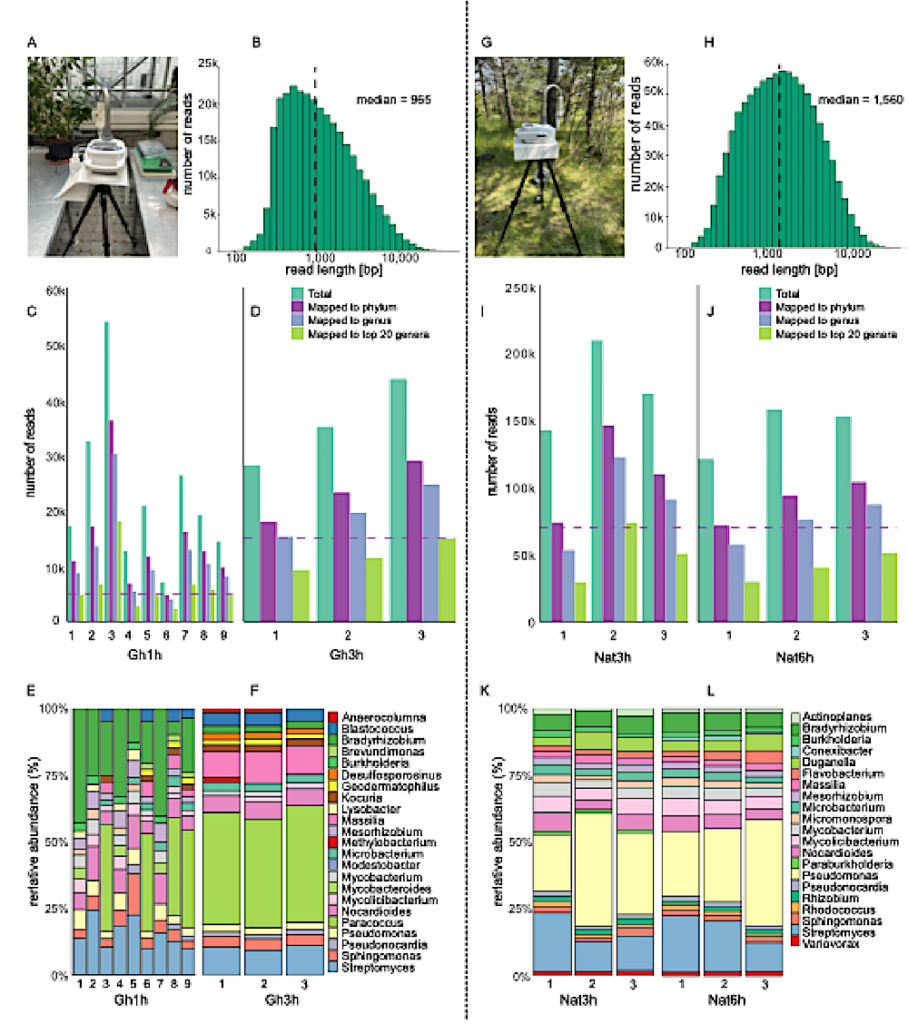
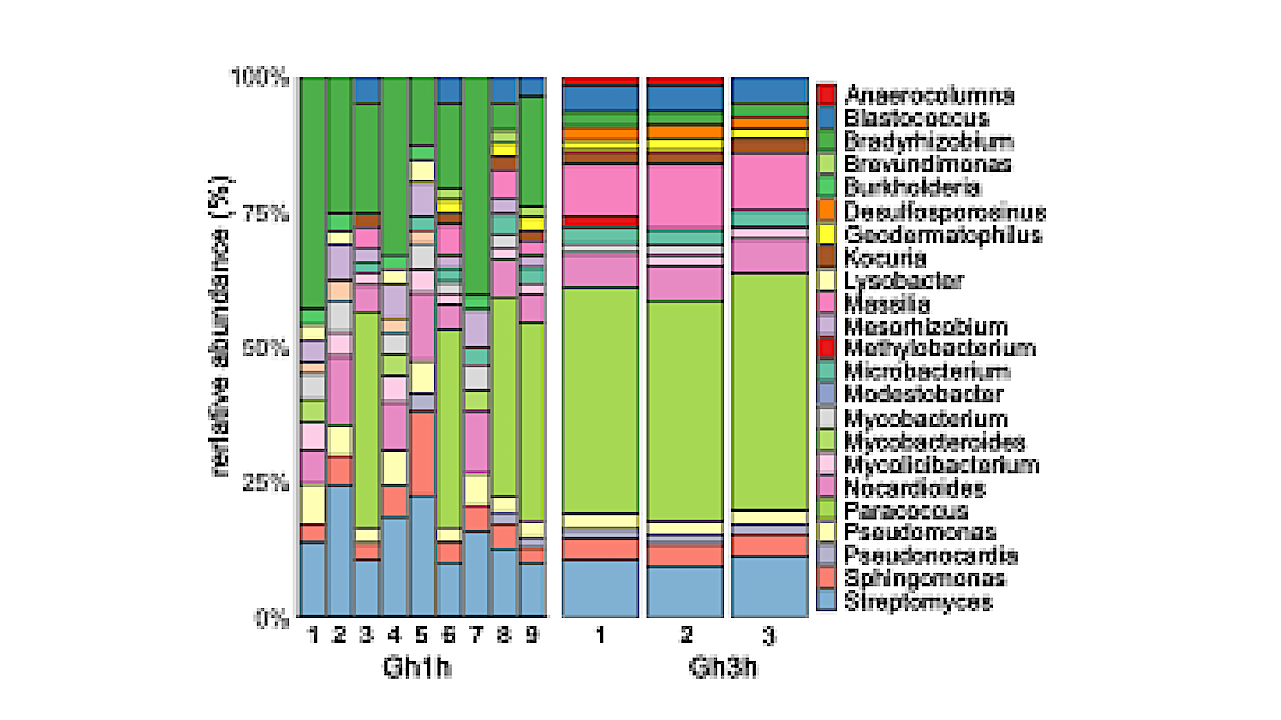
Robust air microbiome assessments through nanopore shotgun sequencing. A. Liquid impingement and nanopore sequencing set-up in a greenhouse (Gh) as controlled environment. B. Nanopore sequencing read length distribution across 1h- and 3-Gh samples. C-D. Number of total sequencing reads, and of reads mapping to taxonomic phylum and genus level as well as to the 20 most dominant genera using Kraken2 (Material and methods) across the C. 1h- and D. 3-Gh samples; the downsampling threshold across samples is indicated by the dashed horizontal line. E-F. Taxonomic composition of the E. 1hand F. 3h-samples after downsampling based on the 20 most dominant genera across samples. G. Air microbiome monitoring in a natural environment (Nat). H. Nanopore sequencing read length distribution across 3h- and 6h-Nat samples. I-J. Number of total sequencing reads, and of reads mapping to taxonomic phylum and genus level as well as to the 20 most dominant genera using Kraken2 across the I. 3h- and J. 6-Gh samples; the downsampling threshold across samples is indicated by the dashed horizontal line. K-L. Taxonomic composition of the K. 3h- and L. 6h-samples after downsampling based on the 20 most dominant genera across samples. — biorxiv.org
Air monitoring by nanopore sequencing, biorxiv.org
Astrobiology,