Solving The Mystery Of Left-handed Amino Acids In Primordial RNA Reactions
While humans have an esthetic liking for symmetry in everything, nature prefers asymmetric, single-handed forms when it comes to amino acids—the building blocks of proteins, and, by extension, all things biological.
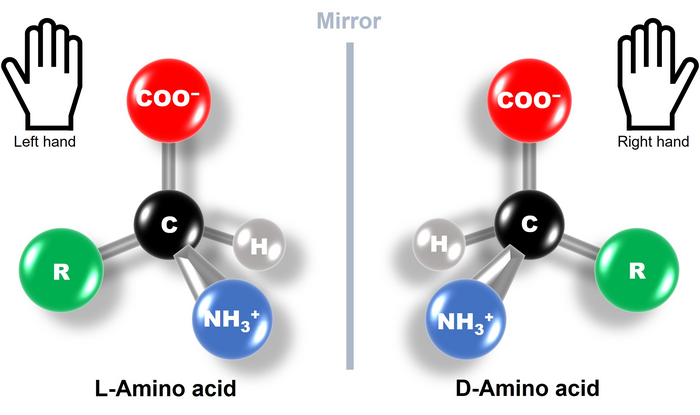
But what does the term left-handed amino acids mean? Amino acids are organic molecules with carbon atoms forming their central skeleton. They usually contain a chiral carbon, i.e., a carbon attached to four different functional groups. This makes amino acids optically active chiral molecules with structures that are mirror images of each other and are not superimposable, just like our left and right hands (Fig. 1). Based on the way these molecules interact and steer the direction of light, they are divided into L-type (left-handed) and D-type (right-handed). What’s fascinating is that all the proteins produced by nature are made up of L-type amino acid chains. This phenomenon is known as biological or amino acid homochirality, and its evolutionary origin and underlying mechanism have been a long-standing scientific puzzle.
Naturally occurring proteins are exclusively left-handed; TUS researchers find out why. CREDIT Tadashi Ando from TUS
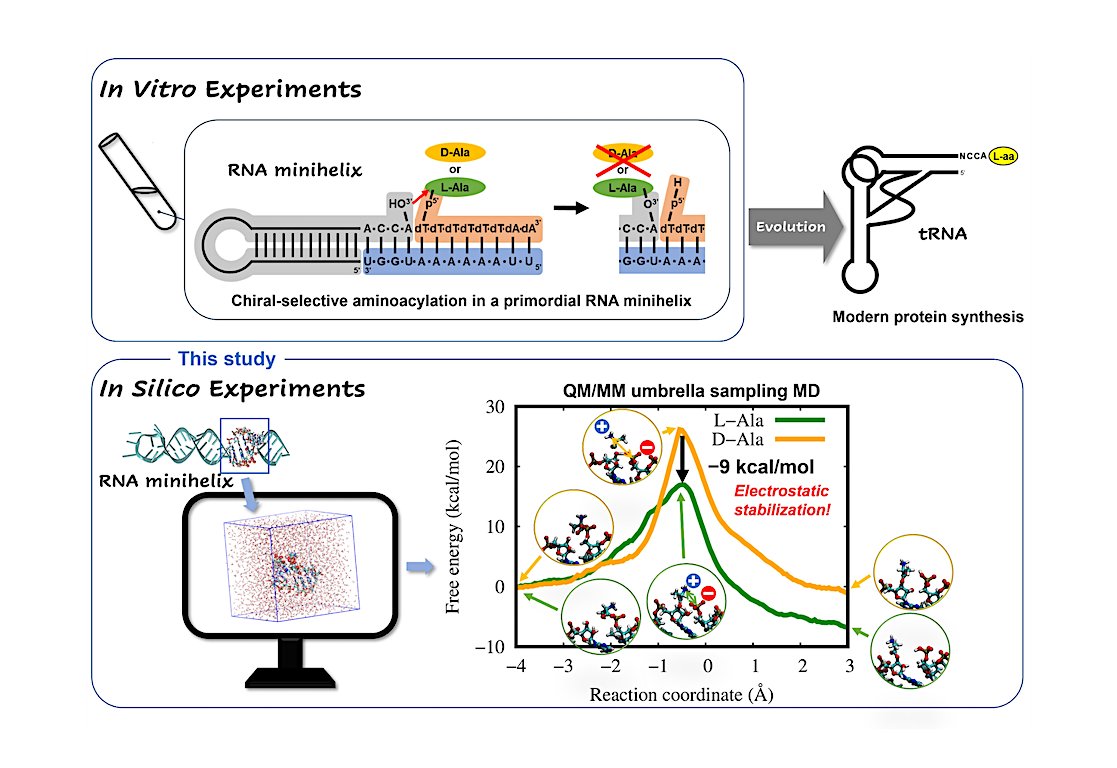
To solve this puzzle, it is important to understand the chiral selectivity seen in certain reactions in primordial RNA minihelices (Fig. 2, upper). So, Associate Professor Tadashi Ando and Professor Koji Tamura from Tokyo University of Science (TUS) investigated the possible mechanisms behind these reactions and their chiral selectivity. Their study published in Life used computer simulations (Fig. 2, lower) to clarify why the amino acid L-alanine was preferred over D-alanine during primordial RNA aminoacylation reactions, without any ribozymes or enzymes steering the selectivity. Aminoacylation reactions are biologically important reactions that involve the attachment of an amino acid to a tRNA during translation. “Many previous studies have suggested that chiral selectivity in aminoacylation might be caused by the steric clash of the amino acid side chain in the constraint of a double helical conformation. However, this has not been fully explained.” stated Dr. Ando while highlighting the fact that “This study will help us get closer to understanding how organisms on earth came to utilize L-amino acids and what role they play in the evolutionary continuum.”
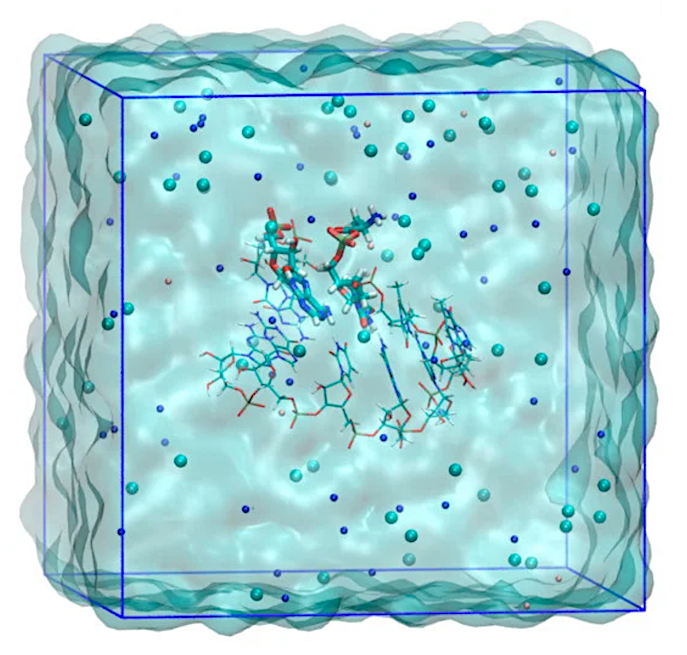
Simulation system for QM/MM umbrella sampling MD calculations for the modeled 6-base pair RNA minihelix. The image is for the D-Ala system. The residues treated with QM calculation are represented by a thick stick model. Atom colors are: oxygen (red), nitrogen (blue), carbon (cyan), phosphorus (brown), and hydrogen (white). The ions of sodium (blue), chloride (cyan) and magnesium (pink) are shown by spheres. Solvent water molecules are rendered as a cyan surface. The image was created using VMD [42].
Direct observation of chiral selective reactions in RNA via experimentation is quite challenging. So, the team adopted a molecular dynamics (MD) simulation strategy that combined the power of quantum mechanical (QM) calculations based on Schrödinger’s equation with molecular mechanics (MM) calculations based on Newton’s classical mechanics. The team went for the QM/MM-based MD method instead of conventional MD methods because the former can provide atomic-level structural pictures at various stages during the chemical reaction, including those involving bond breaking and formation, unlike the latter.
Once equipped with a robust QM/MM strategy, the team ran simulations of L- and D-alanine aminoacylation reactions on modeled RNA. A free-energy profile analysis revealed that the energy barrier for the L-alanine reaction was 9 kcal/mol lower than that of the D-alanine reaction (Fig. 2, lower). The model mechanisms also showed while in its transition state—i.e, a short-lived molecular configuration where the reaction energy is maximum—L-amino acid had much more electrostatic stability than its D-type counterpart, due to the geometrical arrangement of functional groups in its structure. This observation provided a plausible reason for the previously unexplained trend of selective aminoacylation of L-amino acids in RNA minihelices.
The insights presented in the paper have brought us a step closer to understanding the mechanisms driving the homochirality of amino acids, which is an essential step toward decoding the chemical origin of life. The team believes that in addition to unraveling this mystery, the precise use of QM/MM calculations highlighted in the study will encourage more researchers to use these techniques for further fundamental and application-based research. “tRNA aminoacylation is a key reaction in protein synthesis, and the elucidation of L-amino acid selectivity using computational science is expected to lead to new developments in protein engineering and nucleic acid engineering that control chiral selective reactions” concludes Dr. Ando.
Researchers at TUS are unlocking the universal preference one reaction at a time!
Mechanism of Chiral-Selective Aminoacylation of an RNA Minihelix Explored by QM/MM Free-Energy Simulations, Life (open access)
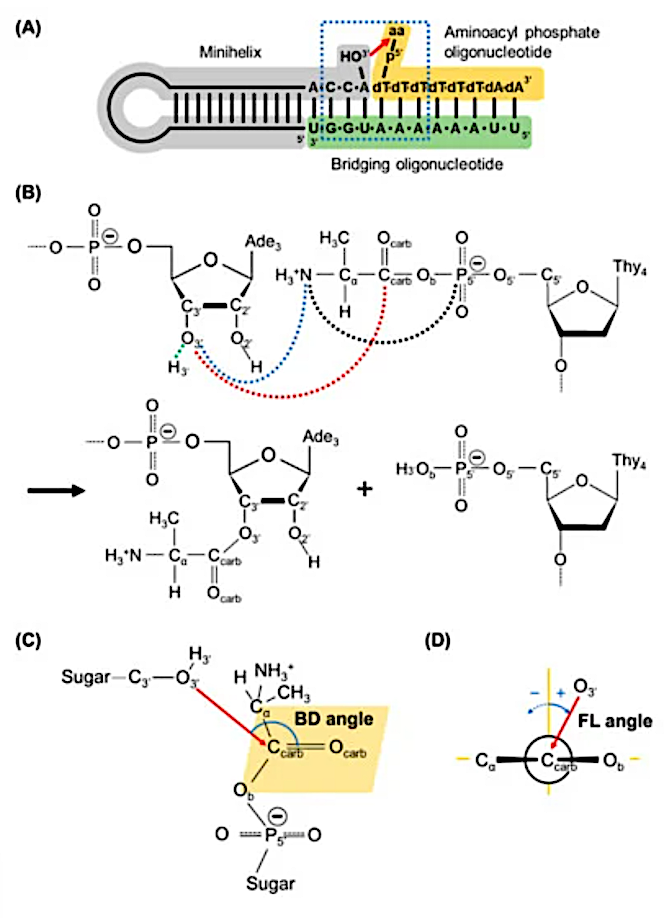
Schematic view of aminoacylation reaction in an RNA minihelix. An aminoacyl moiety is transferred from 5′-phosphate of deoxythymine (dT) of the oligonucleotide to the 3′-hydroxyl group of adenosine (A) of the minihelix. Here aa represents amino acid. The 6-base pair nucleotide sequences surrounded by the blue-dotted rectangle with aa of alanine were modeled in this simulation study. (B) A reaction scheme for the aminoacylation found in the RNA minihelix. Distances between carbonyl carbon (Ccarb) and 3′-hydroxyl oxygen (O3′) atoms, d (Ccarb…O3′), and between 3′-hydrogen atom (H3′) and O3′, d (H3′ …O3′) are drawn with red- and green-dotted lines, respectively, which are used for defining the reaction coordinate for umbrella sampling. The distance between the nitrogen atom of alanine (N) and O3′, d (N…O3′), and between N and the phosphorus atom of 5′-phosphate group (P5′) are drawn with blue- and black-dotted lines, respectively. The abbreviations of atoms used in this study are also described. (C) The Bürgi–Dunitz (BD) angle is defined as the O3′…Ccarb=Ocarb angle in the model. (D) The Flippin–Lodge (FL) angle, defined as the angle between the plane formed by O3′, Ccarb and Ocarb atoms and that perpendicular to the carbonyl plane formed by Ccarb, Ocarb, Cα and Ob atoms (shown as the orange parallelogram in (C)), where Ob is the bridging phosphate oxygen atom. In the study, the FL angle has positive values when the plane formed O3′, Ccarb and Ocarb tilts to the Ob atom. — Life
Astrobiology