Multimodal Techniques For Detecting Alien Life Using Assembly Theory And Spectroscopy
Detecting alien life is a difficult task because it’s hard to find signs of life that could apply to any life form. However, complex molecules could be a promising indicator of life and evolution.
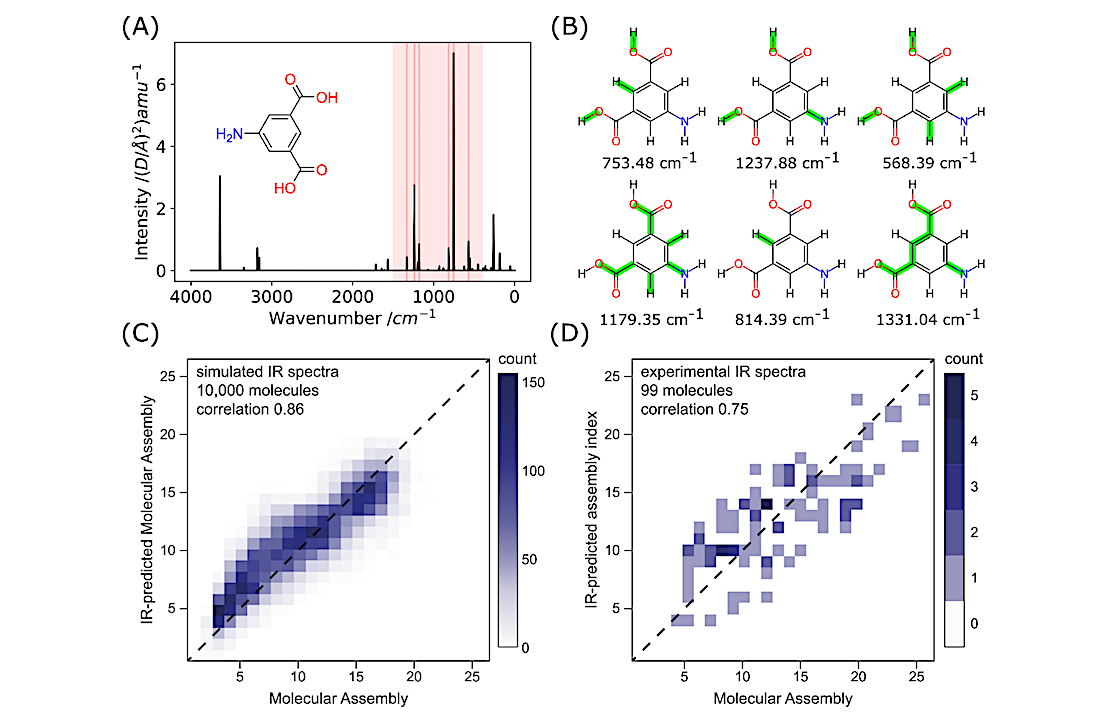
Currently, it’s not possible to experimentally determine how complex a molecule is and how that correlates with information-theoretic approaches that estimate molecular complexity. Assembly Theory has been developed to quantify the complexity of a molecule by finding the shortest path to construct the molecule from simple parts, revealing its molecular assembly index (MA). In this study, we present an approach to rapidly and exhaustively calculate molecular assembly and explore the MA of over 10,000 molecules.
We demonstrate that molecular complexity (MA) can be experimentally measured using three independent techniques: nuclear magnetic resonance (NMR), tandem mass spectrometry (MS), and infrared spectroscopy (IR), and these give consistent results with good correlations. By identifying and counting the number of absorbances in IR spectra, carbon resonances in NMR, or molecular fragments in tandem MS, the molecular assembly index of an unknown molecule can be reliably estimated from experimental data.
This represents the first experimentally quantifiable approach to defining molecular assembly, a reliable metric for complexity, as an intrinsic property of all molecules and can also be performed on complex mixtures.
This paves the way to use spectroscopic techniques to unambiguously detect alien life in the solar system, and beyond on exoplanets.

(a) The general structure of the Go assembly algorithm, with a pool of worker extending pathways. Some features are omitted for brevity, such as branch and bound methods to improve efficiency. (b) A sequence of assembly pathways as processed by the Go algorithm. The top pathway is the starting pathway for the molecule shown, and each subsequent pathway is extended from the pathway above. Pathways are generally extended in multiple ways, and only one such sequence of extensions is shown here. (c) An example of MA values found over time for Primisulfuron-methyl, run to completion, and approximated by stopping early at various stages prior. The new algorithm found pathways at the correct MA of 22 by 10 s, significantly before completion at ~2064 s. The red circle shows split branch algorithm performance on the same molecule. The naïve MA (blue hexagon) is calculated trivially for pathways in which one bond is added at a time (placed illustratively at 10–3 s, as 0 s cannot be represented on the logarithmic scale). — physics.bio-ph
Michael Jirasek, Abhishek Sharma, Jessica R. Bame, Nicola Bell, Stuart M. Marshall, Cole Mathis, Alasdair Macleod, Geoffrey J. T. Cooper, Marcel Swart, Rosa Mollfulleda, Leroy Cronin
Comments: 20 pages, 7 figures, 43 ref
Subjects: Quantitative Methods (q-bio.QM); Biological Physics (physics.bio-ph); Chemical Physics (physics.chem-ph)
Cite as: arXiv:2302.13753 [q-bio.QM] (or arXiv:2302.13753v1 [q-bio.QM] for this version)
https://doi.org/10.48550/arXiv.2302.13753
Focus to learn more
Submission history
From: Leroy Cronin Prof
[v1] Fri, 24 Feb 2023 12:05:57 UTC (5,420 KB)
https://arxiv.org/abs//2302.13753
Astrobiology, Astrochemistry