Immense Diversity And Interdependence In High Temperature Deep-Sea Microorganism Communities

A new study by researchers at Portland State University and the University of Wisconsin finds that a rich diversity of microorganisms live in interdependent communities in high-temperature geothermal environments in the deep sea.
The study, which was published in the journal Microbiome, was led by Anna-Louise Reysenbach, professor of biology at PSU. Emily St. John, who earned a master’s degree in microbial ecology from PSU, also contributed significantly to the study, along with researchers from the University of Wisconsin.
When the 350-400℃ fluid exiting the Earth’s crust through deep-sea hydrothermal vents mixes with sea water it creates large porous rocks often referred to as ‘chimneys’ or hydrothermal deposits. These chimneys are colonized by microbes that thrive in high-temperature environments. For decades, Reysenbach has collected chimneys from deep-sea hydrothermal vents across the world’s oceans, and her lab uses genetic fingerprinting and cultivation techniques to study the microbial diversity of the communities associated with these rocks.
In this new study, Reysenbach and the team were able to take advantage of advances in molecular biology techniques to sequence the entire genomes of the microbes in these communities to learn more about their diversity and interconnected ecosystems.
The team constructed genomes of 3,635 Bacteria and Archaea from 40 different rock communities. The amount of diversity was staggering and greatly expands what is known about how many different types of Bacteria and Archaea exist. The researchers discovered at least 500 new genera (the level of taxonomic organization above species) and have evidence for two new phyla (five levels up from species). “Phyla is way up on the taxonomic rank, so that’s really cool,” says Reysenbach.
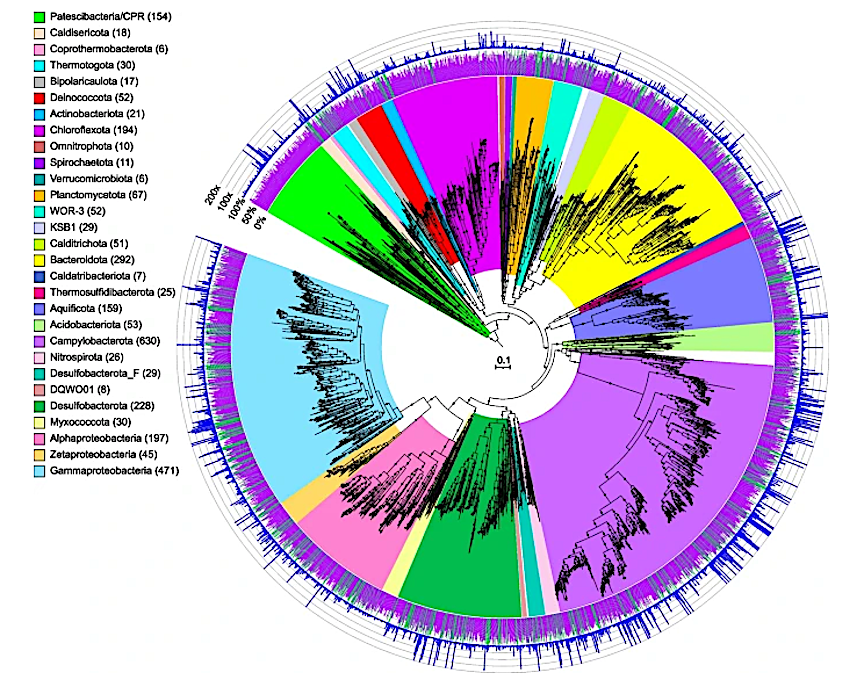
Maximum-likelihood phylogenomic tree of bacterial metagenome-assembled genomes, constructed using 120 bacterial marker genes in GTDB-Tk. Major taxonomic groups are highlighted, and the number of MAGs in each taxon is shown in parentheses. See Table S2 for details. Bacterial lineages are shown at the phylum classification, except for the Proteobacteria which are split into their component classes. The inner ring displays quality (green: high quality, > 90% completion, < 5% contamination; purple: medium quality, ≥ 50% completion, ≤ 10% contamination), while the outer ring shows normalized read coverage up to 200x. The scale bar indicates 0.1 amino acid substitutions per site, and filled circles are shown for SH-like support values ≥ 80%. The tree was artificially rooted with the Patescibacteria using iTOL. The Newick format tree used to generate this figure is available in Data S4, and the formatted tree is available online at https://itol.embl.de/shared/alrlab
The team also found evidence of microbial diversity hotspots. Samples from the deep-sea Brothers volcano located near New Zealand, for instance, were especially enriched with different kinds of microorganisms, many endemic to the volcano.
“That biodiversity was just so huge, off the charts,” says Reysenbach. “At one volcano there was so much new diversity that we hadn’t seen elsewhere before.” This finding may suggest that the increased complexity of the subsurface rocks of a volcano makes them more likely to house diverse microbial species compared to deep-sea hydrothermal vents.
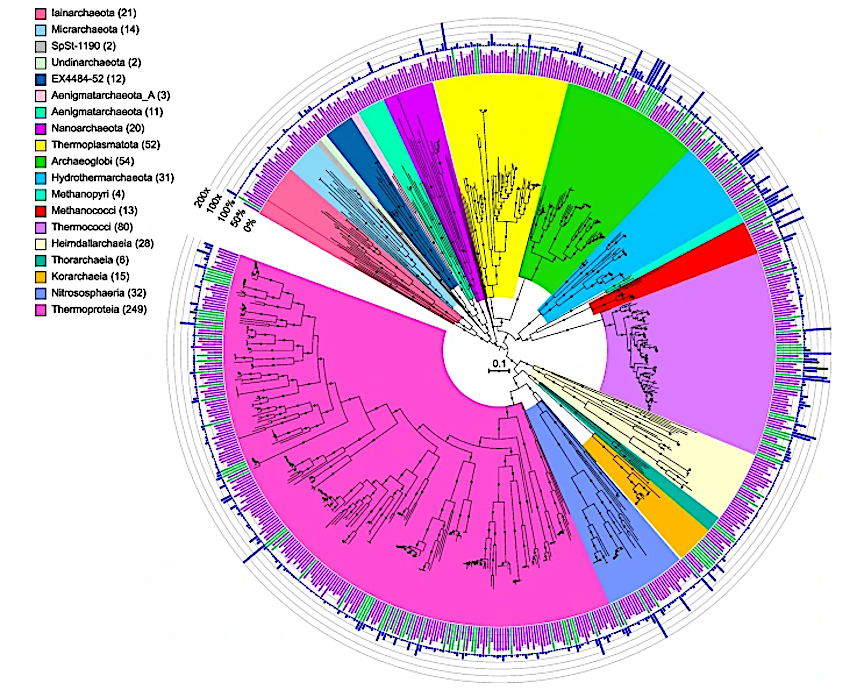
Maximum-likelihood phylogenomic reconstruction of deep-sea hydrothermal vent archaeal metagenome-assembled genomes generated in GTDB-Tk. The tree was generated with 122 archaeal marker genes. Taxa are shown at the phylum level, except for the Thermoproteota, Asgardarchaeota, Halobacteriota, and Methanobacteriota, shown at the class level. The number of MAGs in each highlighted taxon is shown in parentheses. See Table S2 for details. Quality is shown on the inner ring (green: high quality, purple: medium quality, with one manually curated Nanoarchaeota MAG below the 50% completion threshold also displayed as medium quality), while the outer ring displays normalized read coverage up to 200x. SH-like support values ≥ 80% are indicated with filled circles, and the scale bar represents 0.1 amino acid substitutions per site. The tree was artificially rooted with the Iainarchaeota, Micrarchaeota, SpSt-1190, Undinarchaeota, Nanohaloarchaeota, EX4484-52, Aenigmarchaeota, Aenigmarchaeota_A, and Nanoarchaeota using iTOL. The tree used to create this figure is available in Newick format (Data S5), and the formatted tree is publicly available on iTOL at https://itol.embl.de/shared/alrlab
Besides finding a jaw-dropping amount of microorganism biodiversity in these high-temperature ecosystems, the genomic data from this study also showed that many of these organisms depend on one another for survival. By analyzing the genomes, the researchers found that some microorganisms cannot metabolize all of the nutrients they need to survive so they rely on nutrients created by other species in a process known as a “metabolic handoff.”
“By looking at these genomes, we really got a much better understanding of what a lot of these microorganisms are doing and how they’re interacting,” says Reysenbach. “They’re communal; they share food with each other.”
This study has inspired a new phase of research for Reysenbach: gaining an in-depth understanding of the interactions between these deep-sea microorganisms. “For me the most exciting part is that I really want to be able to grow these things in the lab,” says Reysenbach. “I want to be able to see [a microorganism] under a microscope and understand if it needs somebody else to live with.”
Global patterns of diversity and metabolism of microbial communities in deep-sea hydrothermal vent deposits, Microbiome (open access)
Astrobiology