Tricorder Tech: NIH Software Assembles Complete Genome Sequences On-demand
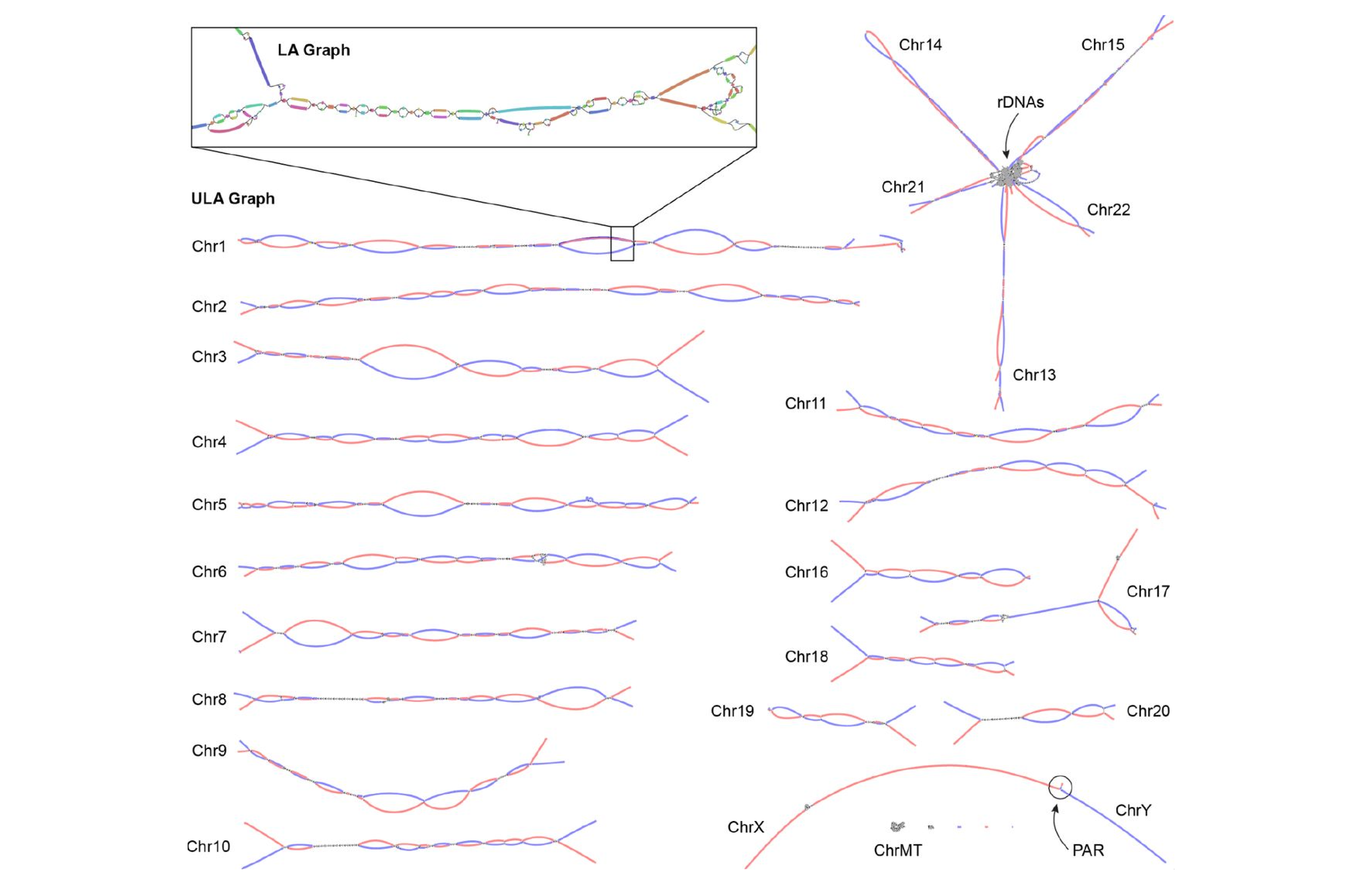
National Institutes of Health researchers have developed and released an innovative software tool to assemble truly complete (i.e., gapless) genome sequences from a variety of species.
This software, called Verkko, which means “network” in Finnish, makes the process of assembling complete genome sequences more affordable and accessible. A description of the new software was published today in Nature Biotechnology.
Verkko grew from assembling the first gapless human genome sequence, which was finished last year by the Telomere-to-Telomere (T2T) consortium, a collaborative project funded by the National Human Genome Research Institute (NHGRI), part of NIH.
“We took everything we learned in the T2T project and automated the process,” said NHGRI associate investigator Sergey Koren, Ph.D., who led the creation of Verkko and is senior author on the paper. “Now with Verkko, we can essentially push a button and automatically get a complete genome sequence.”
The T2T consortium used new DNA sequencing technologies and analytical methods to generate and assemble the remaining 8-10% of the human genome sequence. However, the researchers assembled those fragments manually — a process that took this massive and highly skilled team several years to complete. Verkko can finish the same task in a couple of days.
Assembling a genome sequence is like putting together a jigsaw puzzle, and different DNA sequencing technologies generate different types of genomic puzzle pieces. Some are small and highly detailed, while others are much bigger though the image is blurry. Verkko compares and assembles both types of pieces to generate a complete and accurate picture.
Verkko starts by putting together the small, detailed pieces, creating many partially assembled but disconnected segments of sequence. Then, Verkko compares the assembled regions with the larger, less precise pieces. These larger pieces serve as a framework to order the more detailed regions. The final product is an accurate and complete genome sequence.
The researchers tested Verkko with human and non-human genome sequencing data. The software quickly and precisely assembled the sequences of whole chromosomes, which was once a painstaking feat.
As Verkko leads to more complete human genome sequences, researchers can better assess human genomic diversity. With only one gapless human genome sequence, scientists currently lack knowledge about the diversity of many portions of the genome, such as regions of highly repetitive DNA, across the human population.
Verkko will also accelerate efforts to generate gapless genome sequences of species commonly used in research, such as mice, fruit flies and zebrafish, improving their usefulness to scientists. Additionally, generating gapless genome sequences from a variety of plants, animals and other organisms will aid in comparative genomics, the study of the differences and similarities among the genomes of diverse species.
“Verkko can democratize generating gapless genome sequences,” said Adam Phillippy, Ph.D., an NHGRI senior investigator who worked on the T2T project and the development of Verkko. “This new software will make assembling complete genome sequences as affordable and routine as possible.”
About the National Human Genome Research Institute (NHGRI): At NHGRI, we are focused on advances in genomics research. Building on our leadership role in the initial sequencing of the human genome, we collaborate with the world’s scientific and medical communities to enhance genomic technologies that accelerate breakthroughs and improve lives. By empowering and expanding the field of genomics, we can benefit all of humankind. For more information about NHGRI and its programs, visit www.genome.gov.
About the National Institutes of Health (NIH): NIH, the nation’s medical research agency, includes 27 Institutes and Centers and is a component of the U.S. Department of Health and Human Services. NIH is the primary federal agency conducting and supporting basic, clinical, and translational medical research, and is investigating the causes, treatments, and cures for both common and rare diseases. For more information about NIH and its programs, visit www.nih.gov.
Telomere-to-telomere assembly of diploid chromosomes with Verkko, Nature Biotechnology
Verkko: telomere-to-telomere assembly of diploid chromosomes, Biorxiv
Astrobiology