Host-associated Phages Disperse Across The Extraterrestrial Analogue Antarctica
Extreme Antarctic conditions provide one of the closest analogues of extraterrestrial environments. Since air and snow samples, especially from polar regions, yield DNA amounts in the lower picogram range, binning of prokaryotic genomes is challenging and renders studying the dispersal of biological entities across these environments difficult.
Here, we hypothesized that dispersal of host-associated bacteriophages (adsorbed, replicating or prophages) across the Antarctic continent can be tracked via their genetic signatures aiding our understanding of virus and host dispersal across long distances. Phage genome fragments (PGFs) reconstructed from surface snow metagenomes of three Antarctic stations were assigned to four host genomes, mainly Betaproteobacteria including Ralstonia spp.
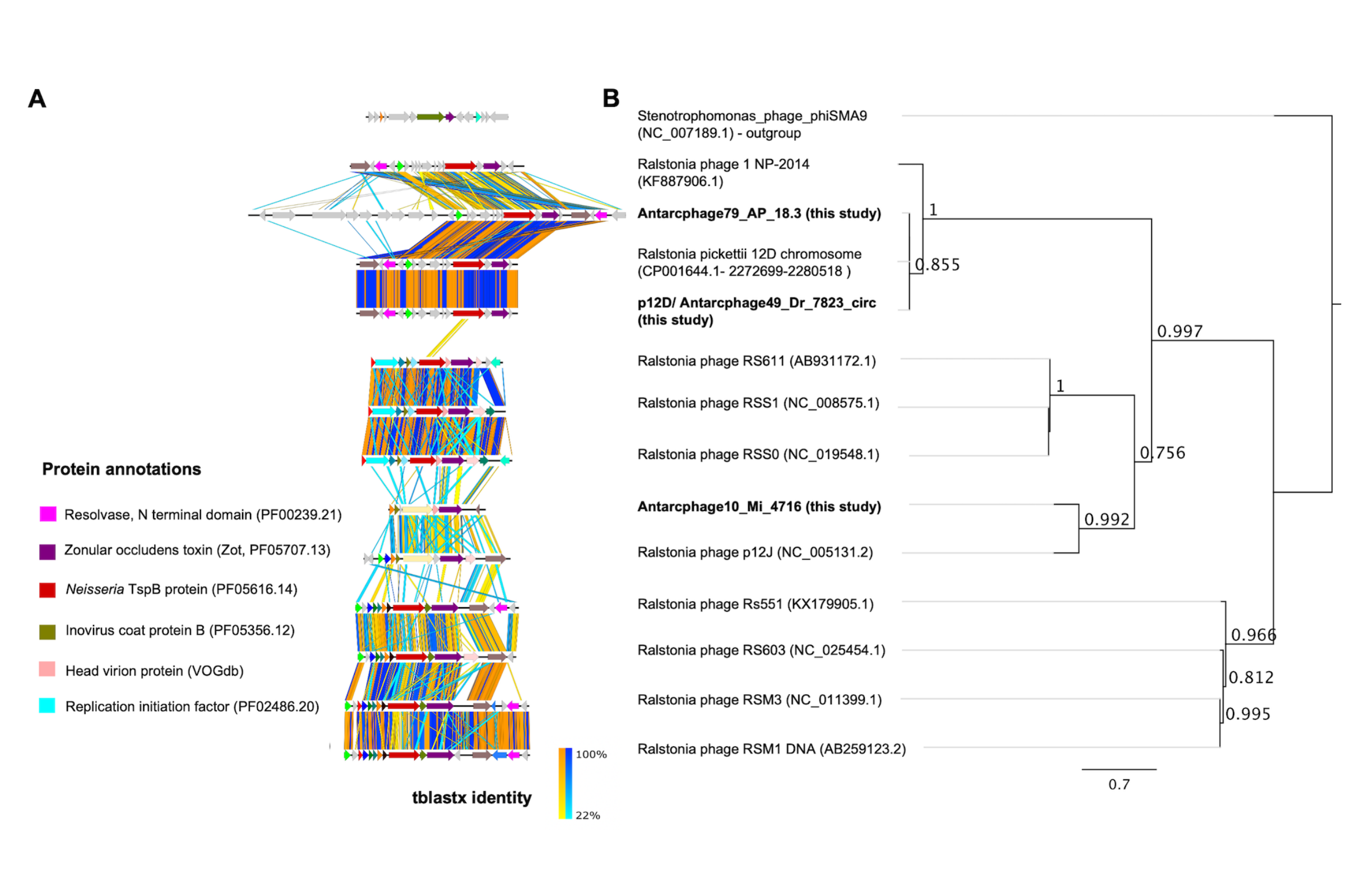
We reconstructed the complete genome of a temperate phage with near-complete alignment to a prophage in the reference genome of Ralstonia pickettii 12D. PGFs from different stations were related to each other at the genus level and matched similar hosts. Metagenomic read mapping and nucleotide polymorphism analysis revealed a wide dispersal of highly identical PGFs, 13 of which were detected in seawater from the Western Antarctic Peninsula in distance of 5538 km to the snow sampling stations.
Our results suggest that host-associated phages, especially of Ralstonia sp., disperse over long distances despite harsh conditions of the Antarctic continent. Given that 14 phages associated with two R. pickettii draft genomes isolated from space equipment were identified, we conclude that Ralstonia phages are ideal mobile genetic elements to track dispersal and contamination in ecosystems relevant for Astrobiology.
Importance Host-associated phages of the bacterium Ralstonia identified in snow samples can be used to track microbial dispersal over thousands of kilometers across the Antarctic continent, which functions as an extraterrestrial analogue because of its harsh environmental conditions. Due to presence of these bacteria carrying genome-integrated prophages on space-related equipment, and the here demonstrated potential for dispersal of host-associated phages, our work has implications for Planetary Protection, a discipline in Astrobiology interested in preventing contamination of celestial bodies with alien biomolecules or forms of life.
Competing Interest Statement
The authors have declared no competing interest.
Janina Rahlff, Till L.V. Bornemann, Anna Lopatina, Konstantin Severinov, Alexander J. Probst
doi: https://doi.org/10.1101/2021.11.09.467789
Astrobiology