Comb Jellies Reveal Ancient Origins Of Animal Genome Regulation
Life depends on genes being switched on and off at exactly the right time. Even the simplest living organisms do this, but usually over short distances across the DNA sequence, with the on/off switch typically right next to a gene. This basic form of genomic regulation is probably as old as life on Earth.
A new study published today in Nature by researchers at the Centre for Genomic Regulation (CRG) and the Centre Nacional d’Anàlisi Genòmica (CNAG) finds that the ability to control genes from far away, over many tens of thousands of DNA letters, evolved between 650 and 700 million years ago. It probably appeared at the very dawn of animal evolution, around 150 million years earlier than previously thought.
Long-distance gene control, called distal regulation, relies on physically folding DNA and proteins into sophisticated loops. This allows regions far from a gene’s starting point to activate its function. This extra layer of control likely helped the first multicellular animals build specialised cell types and tissues without inventing new genes.
The critical innovation likely originated in a sea creature, the common ancestor or all extant animals. The ancient animal evolved the ability to fold DNA in a controlled manner, creating loops in three-dimensional space that brought far-flung bits of DNA in direct contact with each other.
“This creature could repurpose its genetic toolkit in different ways like a Swiss knife, enabling it to refine and explore innovative survival strategies. We did not expect this layer of complexity to be so ancient,” says Dr. Iana Kim, first author of the study and postdoctoral researcher with dual affiliation between the Centre for Genomic Regulation (CRG) and the Centre Nacional d’Anàlisis Genòmica (CNAG).
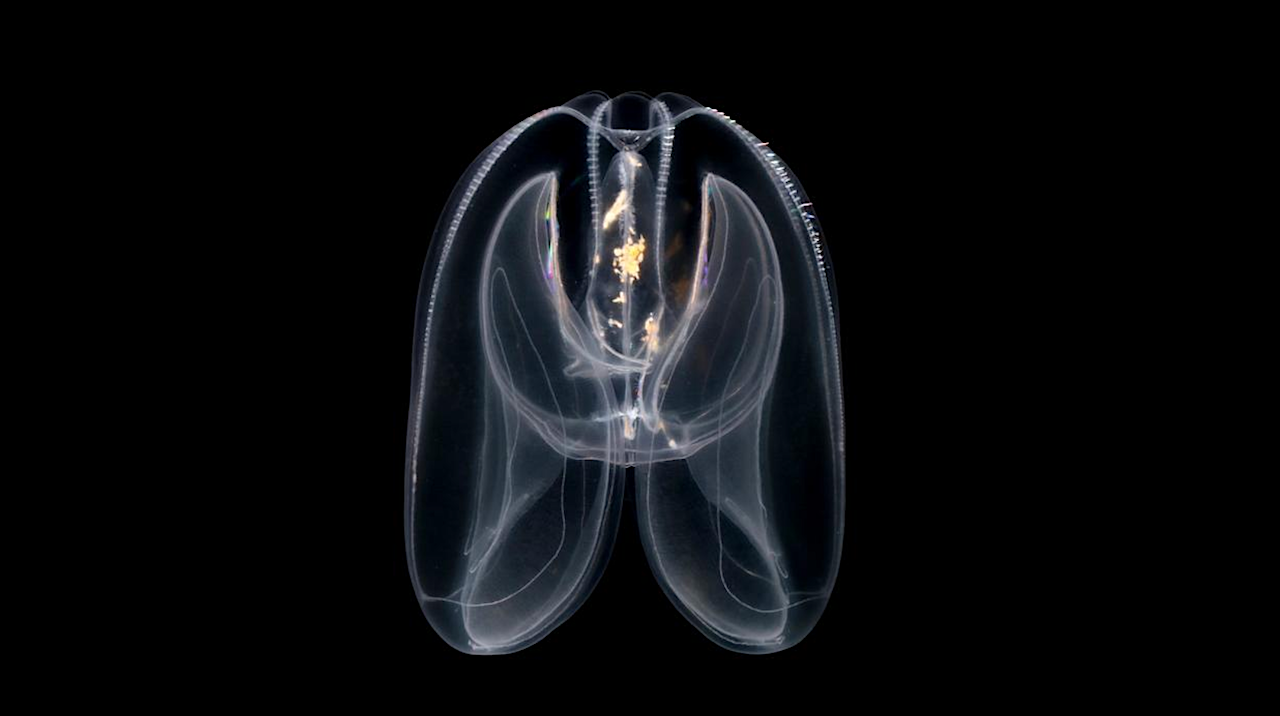
The authors of the study made the discovery by exploring the genomes of many of the oldest branches on the animal family tree, including comb jellies like the ‘sea walnut’ (Mnemiopsis leidyi), placozoans, cnidarians, and sponges. They also studied single-celled relatives that are not animals but share a recent common ancestor.
“You can discover a lot of new biology by looking at weird sea creatures. So far, we had been comparing genome sequences, but thanks to new methods we can now analyse which gene regulation mechanisms control genome function across species,” explains ICREA Research Professor Arnau Sebe-Pedrós, corresponding author of the study and Group Leader at the Centre for Genomic Regulation.
The team used a technique called Micro-C to map how DNA physically folds inside the cells of each of the 11 different species they studied. For scale, each human cell nucleus packs about two metres of DNA. The researchers sifted through 10 billion pieces of sequencing data to build each species’ 3D genome map in detail.

a, Example of syntenic genomic regions in placozoans T. adhaerens (TadhH1_4: 3860000–4060000, bin 800 bp) and C. collaboinventa (chr. 4: 8983000–9183000, bin 800 bp). b, Gaudí plots projecting ATAC-seq, H3K4me3 and exon annotation signals onto a two-dimensional Kamada–Kawai graph layout (top left) represented by the top 20% of contact pairs with solid colours highlighting statistically significant regions (P < 0.05) identified using a one-sided permutation test. The high–high (HH) signal marks genomic bins enriched in signal and that are in spatial proximity with other bins enriched in signal; low–low (LL) bins are depleted in signal as well as neighbourhoods are depleted in signal; high–low (HL) and low–high (LH) are bins that are enriched in signal, but not their neighbourhood, and in reverse. c, Classification of T. adhaerens genes into three categories (GP1, GP2 and GP3) on the basis of structural and epigenetic features. Top, example regions containing genes classified into GP1, GP2 and GP3 groups. The resolution of Micro-C maps is 800 bp, maximum intensity value of ICE normalized Micro-C maps is as in a. Bottom, average loop strength between promoter regions of the genes from each groups is measured with APA. The colour bar of pile-up plots shows enrichment of observed over expected values. d, Sequence motif found in loop regions, which are also overlapping GP1 promoter regions (left panel), is present in promoter regions of orthologous GP1 genes in other placozoan species (right panel). The total number of shared orthologues is indicated. TrH2, Trichoplax sp. H2; Hhon, Hoilungia hongkongensis; HoiH23, C. collaboinventa. e, Heatmaps showing CPM normalized ATAC-seq and ChIP–seq coverage, motif scores and Mutator transposable element density ±5 kb around the TSS of GP1, GP2 and GP3 genes. Each heatmap scale starts at zero. — Nature
While there was no evidence of distal regulation in the single-celled relatives of animals, early-branching animals like comb jellies, placozoans and cnidarians had many loops. The sea walnut alone had over four thousand loops genome-wide. The finding is surprising given its genome is around just 200 million DNA letters long. In comparison, the human genome is 3.1 billion letters long and our cells can have tens of thousands of loops.
Until now, distal regulation was thought to have first appeared in the last common ancestor of bilaterians, a group of many different types of animals which first appeared on Earth around 500 million years ago. However, comb jellies are descended from life forms which diverged early from other animal lineages around 650 to 700 million years ago.
Whether comb jellies are older than sponges in the tree of life is a longstanding debate in evolutionary biology circles, but the study demonstrates that distal regulation arose at least one hundred and fifty million years earlier than previously thought.
The study made another surprising discovery. Many animals are vertebrates. In their cells, loops are controlled by CTCF, an architectural protein which defines boundaries and compartmentalises genes into different local neighbourhoods. It is a foundational unit of genomic architecture in mammals, birds, reptiles, amphibians, and fish. However, the genomes of the early-branching animals do not encode any equivalent protein to CTCF. Instead, the authors discovered that comb jellies use a different architectural protein belonging to the same structural family. The discovery shatters the assumption that advanced genomic, distal regulation require CTCF.
“It is impressive that the same problem has been solved using different tools. Thanks to this work, we now know that you can use two different proteins to bring distal DNA pices together in space forming a loop. Isn’t evolution marvellous?” says ICREA Research Professor Marc A. Marti-Renom, Group Leader at the Centre Nacional d’Anàlisi Genomic and the Centre for Genomic Regulation.
Like sponges and comb jellies, humans are also made of the same building blocks of DNA. Today, our bodies rely on the ancient innovation of distal regulation to help create different types of cells from the same DNA, producing everything from brain cells to immune cells. When these contacts go wrong, diseases can arise.
By tracing distal regulation to animals that lived many hundreds of million years ago, researchers can begin to piece together how the earliest versions of genomic regulation took shape, providing new clues about the fundamental principles that govern our cells and bodies today. This can help us understand where the system is robust and where it’s prone to failure, potentially guiding new medical insights or therapies.
This work was led by the Centre for Genomic Regulation (CRG), in collaboration with Marc A. Marti Renom’s Group at the Centre Nacional d’Anàlisis Genòmica, the University of Bergen, Queen Mary University of London, the Prefectural University of Hiroshima, and the University of Alberta. It was funded by an ERC Starting Grant (European Research Council) from the European Union.
Chromatin loops are an ancestral hallmark of the animal regulatory genome, Nature (open access)
Astrobiology, Genomics, Evolution,