Structural Evolution Of Nitrogenase Enzymes Over Geologic Time
Life on Earth is more than 3.5 billion years old, nearly as old as the age of the planet. Over this vast expanse of time, life and its biomolecules adapted to and triggered profound changes to the Earth s environment.
Certain critical enzymes evolved early in the history of life and have persisted through planetary extremes. While sequence data is widely used to trace evolutionary trajectories, enzyme structure remains an underexplored resource for understanding how proteins evolve over long timescales.
Here, we implement an integrated approach to study nitrogenase, an ancient, globally critical enzyme essential for nitrogen fixation. Despite the ecological diversity of its host microbes, nitrogenase has strict functional limitations, including extreme oxygen sensitivity, energy requirements and substrate availability.
We combined phylogenetics, ancestral sequence reconstruction, protein crystallography and deep-learning based structural prediction, and resurrected three billion years of nitrogenase structural history.
We present the first effort to predict all extant and ancestral structures along the evolutionary tree of an enzyme and present a total of over 5000 structures.
This approach lays the foundation for reconstructing key structural constraints that influence protein evolution and examining ancient enzymes in the context of phylogenetic and environmental change across geological timescales.
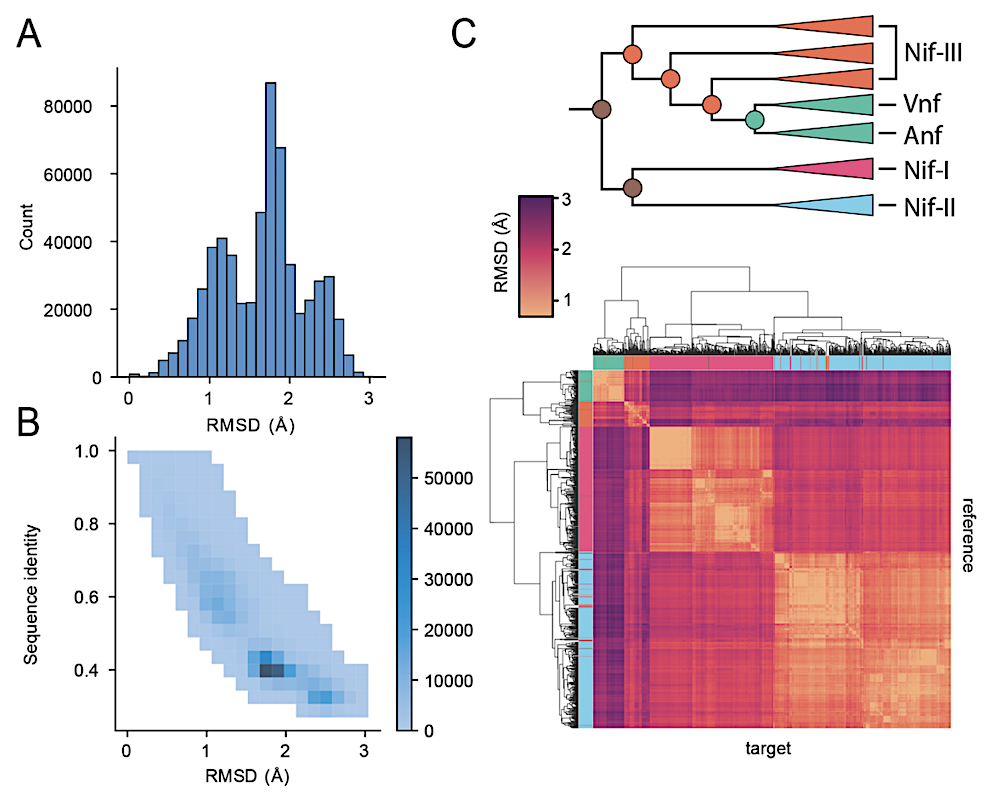
Global analyses of nitrogenase DDKK sequence and structural diversity. A) Root Mean Squared Deviation (RMSD) distribution on paired nitrogenase alignments across extant and ML-ancestor pairs. B) Sequence identity and structural similarity (quantified by root mean square deviation (RMSD) of aligned predicted structures) distribution of paired nitrogenase alignments. C) Hierarchical clustering of predicted nitrogenase structures based on structural similarity (RMSD). Each tile in the heatmap corresponds to the RMSD between two nitrogenase structures. — biorxiv.org

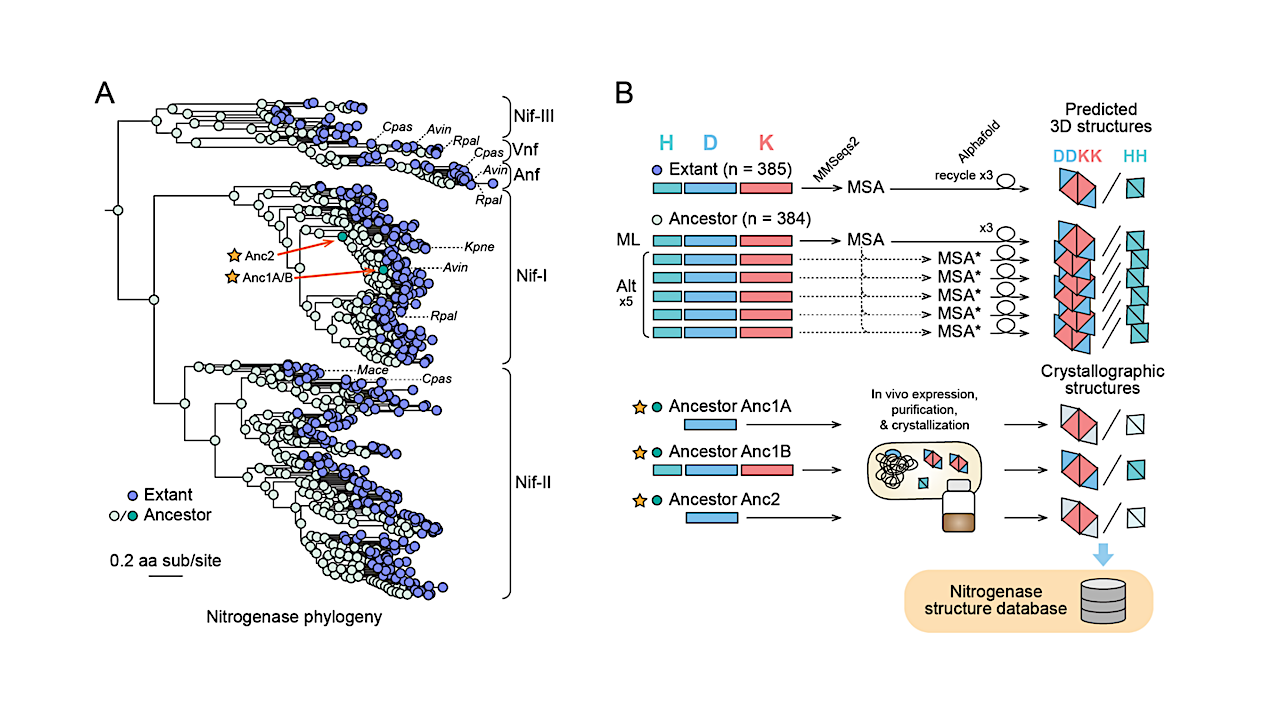
Figure 5. Crystal structures of targeted ancestral nitrogenases. A) Nitrogenase protein phylogeny from which nitrogenase ancestral proteins were reconstructed and crystallized for structural characterization. Major clades are labeled following (Garcia et al., 2023). B) Residue-level RMSD between the crystallized ancestral and wild-type (A. vinelandii) NifDK/NifH structures. C) Spatial distribution of ancestral amino acid substitutions relative to WT. Crystallized protein complexes for Anc1A and Anc2 contain ancestral NifD and WT NifH and NifK. Therefore, ancestral substitutions are only in the NifD subunit for these structures. D) Ancestral amino acid substitutions within the NifH-NifDK interface of Anc1B. available under aCC-BY-NC-ND 4.0 International license. was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made bioRxiv preprint doi: https://doi.org/10.1101/2024.11.18.623660; this version posted April 10, 2025. The copyright holder for this preprint (which 19 Bound positions of NifH are inferred by alignment with either the nucleotide-free (PDB 2AFH (Tezcan et al., 2005)) or MgATP-bound (PDB 7UT8 (Rutledge et al., 2022)) structures. E-G) Close views of specific, ancestral amino acid substitutions that are inferred to impact NifH418 NifDK interactions. — biorxiv.org
Structural evolution of nitrogenase enzymes over geologic time, biorxiv.org
Astrobiology, evolution, genomics,