Tricorder Tech: Unlocking the Full Potential of Nanopore Sequencing: Tips, Tricks, and Advanced Data Analysis Techniques

Editor’s note: If we aspire to mount expeditions to new worlds and then embrace the task of characterizing and quantifying whatever life forms we find, the ability to map and understand whatever metabolic and genomic systems are in operation is important. Not only do we need to know how alien biota function, but also how they evolved – what differences and similarities they may have with the origin and evolution of life on Earth. Increasing in situ capabilities like this can allow much more preliminary analysis to be done on site – or back on Earth.
As we begin to expand our search for life to other worlds we are going to need to be economical interms of the equipment we send and how we reality new knowledge back to Earth. Sample return missions are difficult even when worlds are close to one another. Doing in situ examination and documentation is going to be very important as we explore other worlds. Not only does it reduce the logistics of sending things back home but it allows data to be sent back at the speed of light. It also allows the astronaut/droid teams to engage in empirical exploration – learning from what they found so as to refine and perfect their continued searching.
Nucleic acid sequencing is the process of identifying the sequence of DNA or RNA, with DNA used for genomes and RNA for transcriptomes.
Deciphering this information has the potential to greatly advance our understanding of genomic features and cellular functions. In comparison to other available sequencing methods, nanopore sequencing stands out due to its unique advantages of processing long nucleic acid strands in real time, within a small portable device, enabling the rapid analysis of samples in diverse settings.
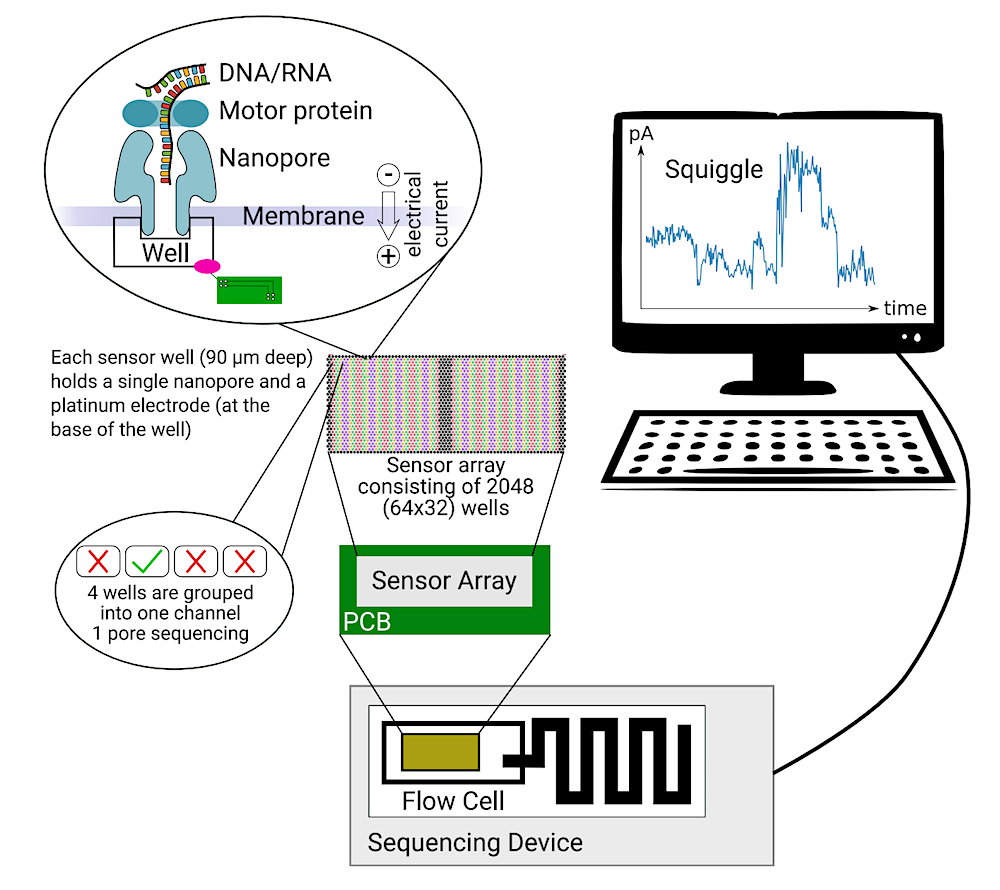
Nanopore sequencing on a MinION. A MinION flow cell contains 512 channels with 4 nanopores in each channel, resulting in 2 048 nanopores available for sequencing DNA or RNA. Each sensor well (90 µm deep) holds a single nanopore and a platinum electrode (at the base of the well). The electrode is controlled via the PCB (Printed Circuit Board) and measures the ion flow as ionic current. During sequencing, a motor protein guides the DNA/RNA through the nanopore following the electric gradient. The DNA/RNA disrupts the current by partially blocking the ion flow. This can be measured in a resulting squiggle.– biorxiv.org
Evolving over the past decade, nanopore sequencing remains in a state of ongoing development and refinement, resulting in persistent challenges in protocols and technology. This article employs an interdisciplinary approach, evaluating experimental and computational methods to address critical gaps in our understanding in order to maximize the information gain from this advancing technology.

Former NASA astronaut Peggy Whitson works on the Genes In Space-3 experiment demonstrating ways in which portable, real-time DNA sequencing can be used to monitor crew health aboard the space station. — NASA
- Timeline Of DNA Sequencing Technology On The International Space Station, Astrobiology.com
- Genomics, Proteomics, Bioinformatics, Astrobiology.com
- MinION postings, Astrobiology.com
Here we present both overview and analysis of all aspects of nanopore sequencing by providing statistically supported insights. Thus, we aim to provide fresh perspectives on nanopore sequencing and give comprehensive guidelines for the diverse challenges that frequently impede optimal experimental outcomes.

TIPS & TRICKS mentioned within the manuscript are summarized here for a better overview. biorxiv.org
Unlocking the Full Potential of Nanopore Sequencing: Tips, Tricks, and Advanced Data Analysis Techniques (open access)
Astrobiology, Genomics,