Unveiling Hidden Microbial Diversity in Mars 2020 Mission Assembly Cleanrooms with Molecular Insights into the Persistent and Perseverance of Novel Species Defying Metagenome Sequencing
Due to strict cleaning procedures and nutrient-scarce conditions, aerospace industrial cleanrooms – which are essential for assembling space mission components – contain low microbial burdens but are tough to eradicate.
Owing to their resilience, persistence, scarcity, and the limitations of existing DNA extraction and sequencing techniques, uncommon extremophiles remain challenging to detect even with state-of-the-art technologies. Rare species are generally neglected by traditional metagenomic approaches because they often do not break down hard microbial cells or capture the few amounts of DNA present.
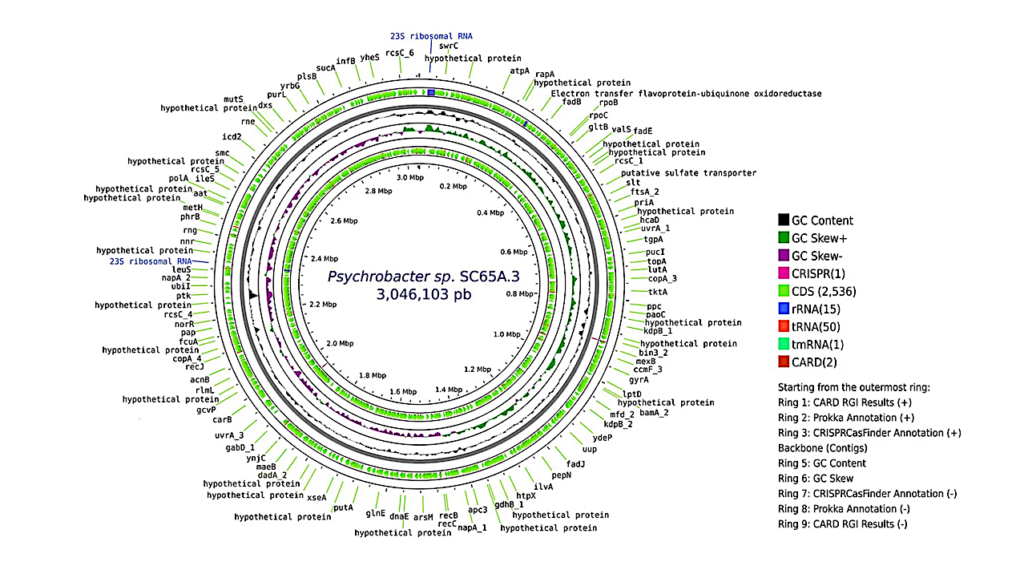
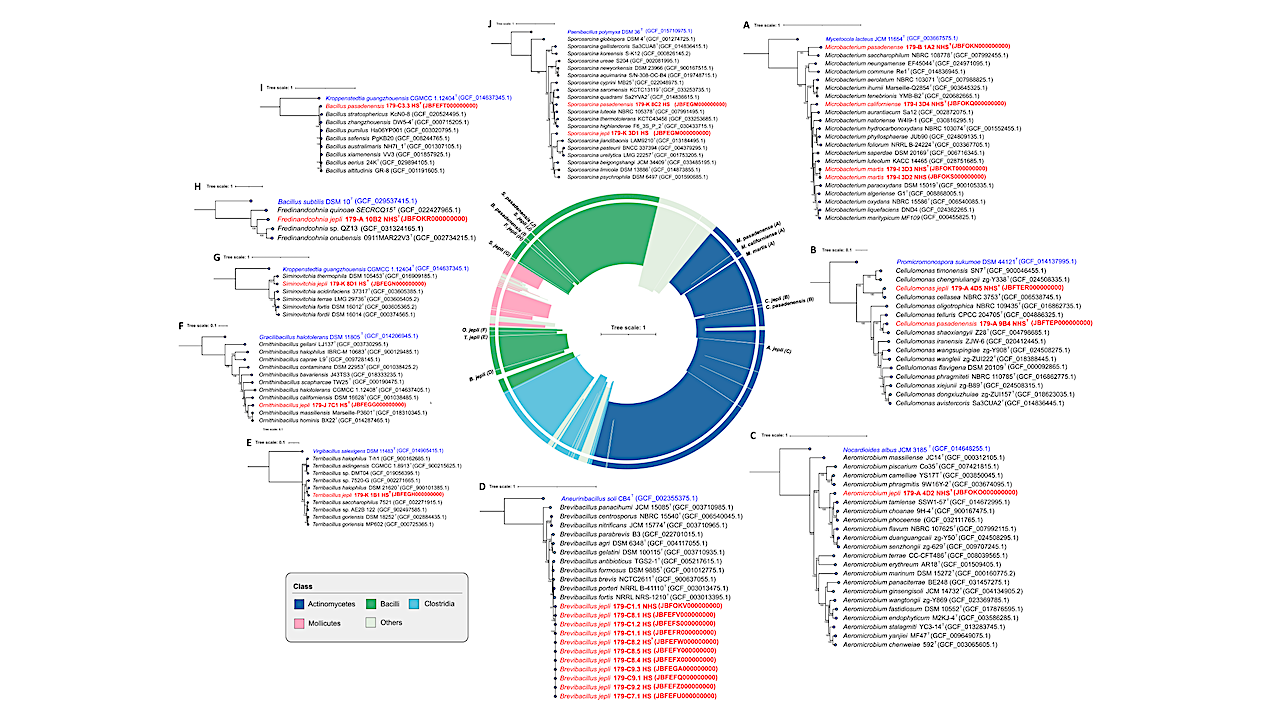
Furthermore, newly identified extremophiles may not have references in the databases currently in use, making identification difficult. During six months of microbiological monitoring at the Mars 2020 mission construction cleanrooms, 182 bacterial strains from 19 families were identified. Fourteen novel Gram-positive species were isolated, including eight spore-forming species.

Overview of the study design including I. Sampling and sequencing procedures, II. Taxonomic classification, III. Mapping of metagenomic reads to estimate the abundance of novel species, and IV. Functional characterization. — biorxiv.org
However, the genomic approach indicates that these microbes exhibit little genetic resemblance to any recognized species, with average nucleotide identity (ANI) values ranging from 77% to 94%. The bacteria were successfully cultured despite their abundance in the metagenome sequencing data being ∼0.001%. The functional characterization of these extremophiles showed that they could cycle nitrogen, break down carbohydrates, and survive radiation.
Potential uses in biotechnology were indicated by the identification of 12 biosynthetic gene clusters, including those associated with the production of ectoine and ϵ-poly-L-lysine. These findings highlight the hidden diversity of cleanrooms and the need for more sophisticated detection techniques to find extremophiles that may help guide future biotechnological advancements and space exploration.
Astrobiology