Tricorder Tech: New AI Technique Generates Clear Images Of Thick Biological Samples Without The Fancy Hardware
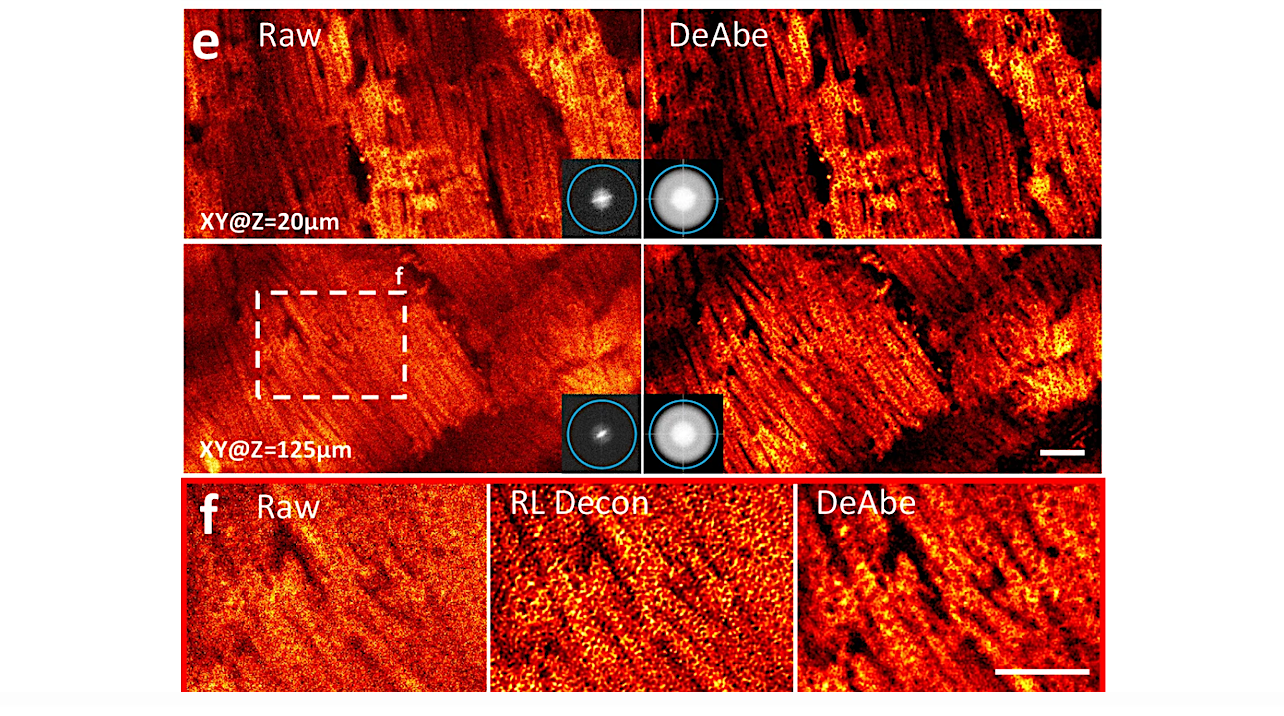
Depth degradation is a problem biologists know all too well: The deeper you look into a sample, the fuzzier the image becomes. A worm embryo or a piece of tissue may only be tens of microns thick, but the bending of light causes microscopy images to lose their sharpness as the instruments peer beyond the top layer.
To deal with this problem, microscopists add technology to existing microscopes to cancel out these distortions. But this technique, called adaptive optics, requires time, money, and expertise, making it available to relatively few biology labs.
Now, researchers at HHMI’s Janelia Research Campus and collaborators have developed a way to make a similar correction, but without using adaptive optics, adding additional hardware, or taking more images. A team from the Shroff Lab has developed a new AI method that produces sharp microscopy images throughout a thick biological sample.
To create the new technique, the team first figured out a way to model how the image was being degraded as the microscope imaged deeper into a uniform sample. They then applied their model to near-side images of the same sample that weren’t degraded, causing these clear images to become distorted like the deeper images. Then, they trained a neural network to reverse the distortion for the entire sample, resulting in a clear image throughout the entire depth of the sample.
Not only does the method produce better looking images, but it also enabled the team to count the number of cells in worm embryos more accurately, trace vessels and tracts in the whole mouse embryos, and examine mitochondria in pieces of mice livers and hearts.
The new deep learning-based method does not require any equipment beyond a standard microscope, a computer with a graphics card and a short tutorial on how to run the computer code, making it more accessible than traditional adaptive optics techniques.
The Shroff Lab is already using the new technique to image worm embryos, and the team plans to further develop the model to make it less dependent on the structure of the sample so the new method can be applied to less uniform samples.
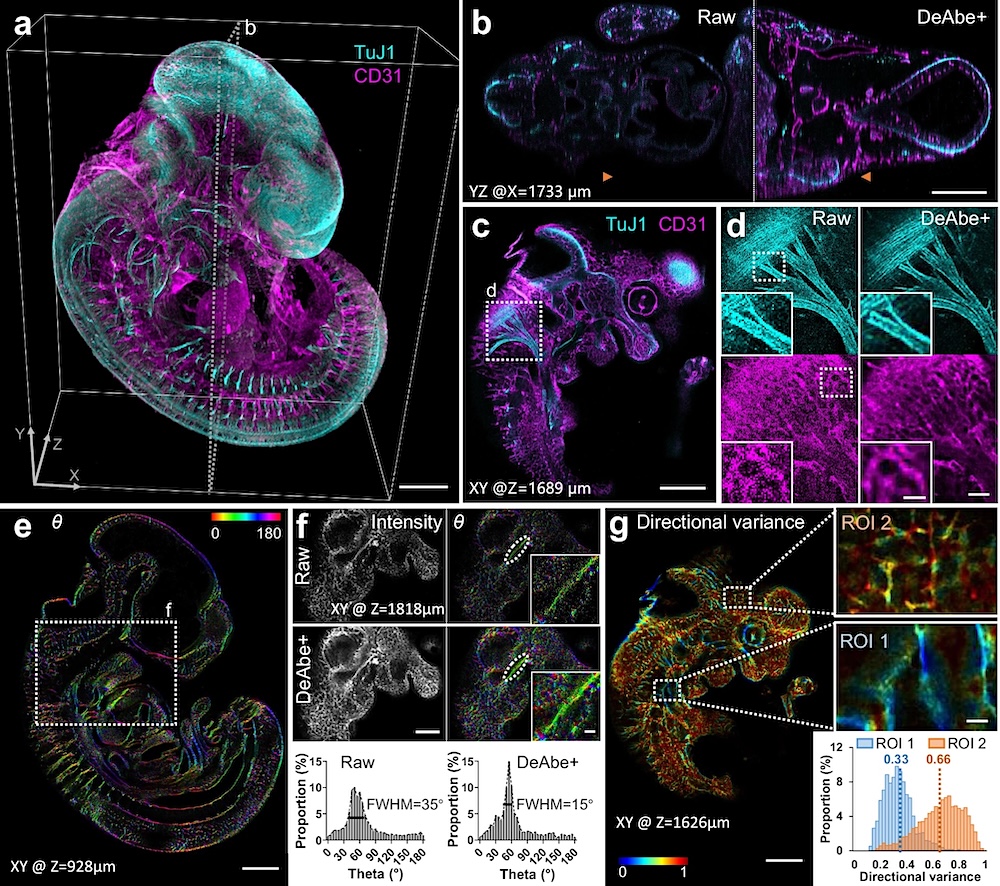
a Fixed and iDISCO-cleared E11.5-day mouse embryos were immunostained for neurons (TuJ1, cyan) and blood vessels (CD31, magenta), imaged with confocal microscopy and processed with a trained DeAbe model. See also Supplementary Movie 8. b Axial view corresponding to dotted rectangular region in (a), comparing raw data and depth-compensated, de-aberrated, and deconvolved data (DeAbe + ). See also Supplementary Figs. 23, 24. c Higher magnification lateral view at axial depth of 1689 μm indicated by the orange double headed arrowheads in (b). d Higher magnification views of white dotted region in (c), comparing raw (left) and DeAbe+ processing (right) for neuronal (top) and blood vessel (bottom) stains. e Orientation (θ, transverse angle) analysis on blood vessel channel of DeAbe+ data, here shown on single lateral plane at indicated axial depth. See also Supplementary Fig. 25, Supplementary Movie 9. f Higher magnification lateral view of white dotted region in (e) (note that axial plane is different), comparing intensity (left) and orientation (right) views between raw (top row) and DeAbe+ prediction (middle row). Righthand insets show higher magnification views of vessel and surrounding region highlighted by dotted lines. Bottom row indicates histogram of all orientations in the vessel highlighted with dotted ellipse, full-width-at-half maximum (FWHM) in peak region of histogram is also shown. g Directional variance of blood vessel stain within the indicated plane, with higher magnification region of interest (ROI) views at right. Histogram of directional variance in both regions also shown. See also Supplementary Fig. 26. Scale bars: 500 μm (a, b, c, e); 100 μm (d), 50 μm inset; 300 μm (f), 50 μm inset; 300 μm (g), 50 μm inset. Data shown are representative samples from N = 3 experiments for (a–d) and N = 1 for(e–g). — Nature Communications
Deep learning-based aberration compensation improves contrast and resolution in fluorescence microscopy, Nature Communications (open access)
Astrobiology








