NASA Open Science Data Repository: Open Science For Life In Space

Space biology and health data are critical for the success of deep space missions and sustainable human presence off-world. At the core of effectively managing biomedical risks is the commitment to open science principles, which ensure that data are findable, accessible, interoperable, reusable, reproducible and maximally open.
The 2021 integration of the Ames Life Sciences Data Archive with GeneLab to establish the NASA Open Science Data Repository significantly enhanced access to a wide range of life sciences, biomedical-clinical and mission telemetry data alongside existing ‘omics data from GeneLab.
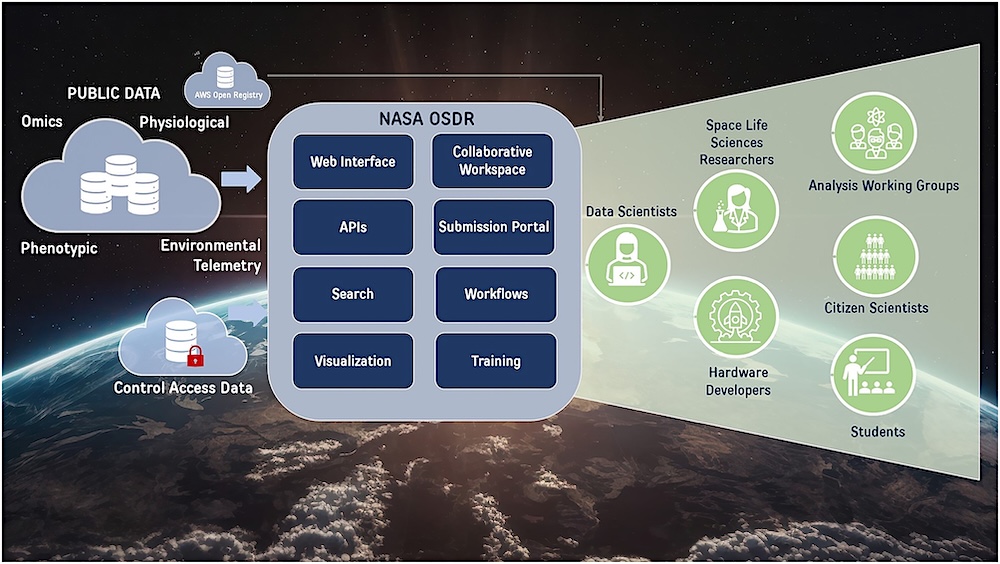
This paper describes the new database, its architecture and new data streams supporting diverse data types and enhancing data submission, retrieval and analysis. Features include the biological data management environment for improved data submission, a new user interface, controlled data access, an enhanced API and comprehensive public visualization tools for environmental telemetry, radiation dosimetry data and ‘omics analyses.
By fostering global collaboration through its analysis working groups and training programs, the open science data repository promotes widespread engagement in space biology, ensuring transparency and inclusivity in research.
It supports the global scientific community in advancing our understanding of spaceflight’s impact on biological systems, ensuring humans will thrive in future deep space missions.
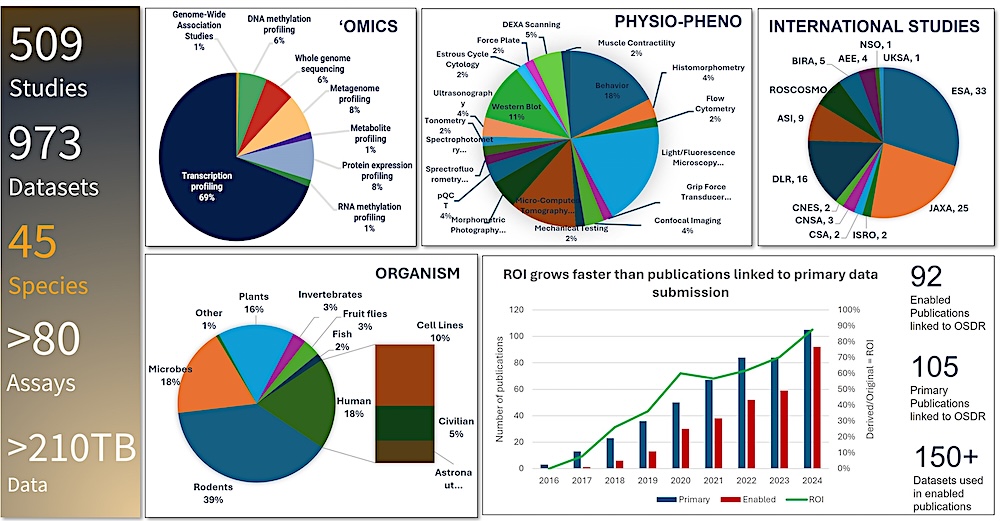
OSDR metrics. Overview of studies hosted on OSDR as of October 2024, including the total number of international studies, percent of total datasets for each ‘omics and each phenotypic-physiological measurement type, percent of total studies for each organism and the ROI measured by the number of enabled publications versus primary publications that use OSDR datasets. — NASA
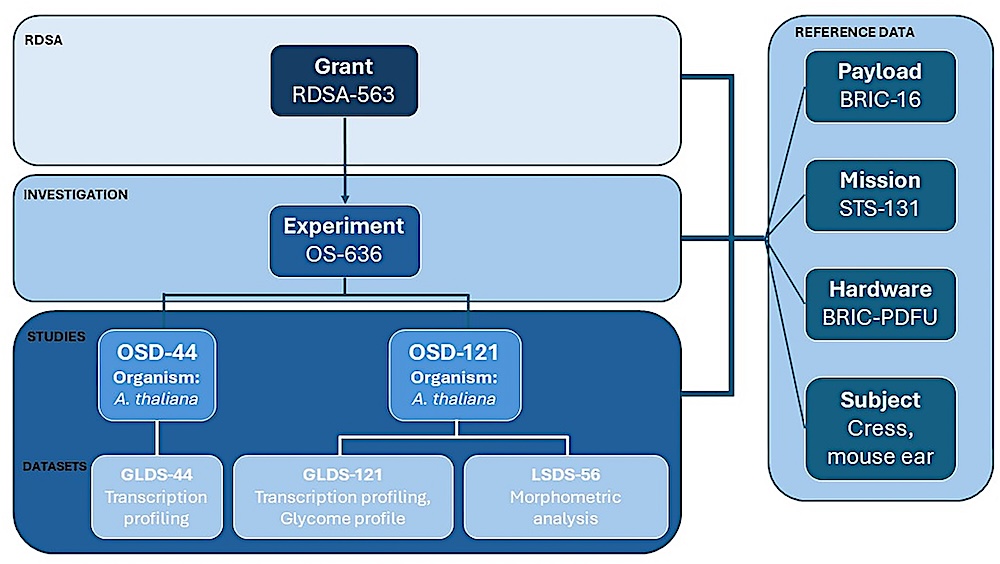
NASA funded PI data and metadata hierarchy. The RDSA is for NASA-funded PIs and describes the experiment(s) conducted with the associated grant funds. Each experiment receives a unique identifier (e.g. OS-636) and is linked to respective Reference data and studies. Reference data records associated with each experiment include payload (e.g. BRIC-16), mission (e.g. STS-131), hardware (e.g. BRIC-PDFU) and subject (e.g. Cress, mouse ear). Each study associated with an experiment has a unique identifier (e.g. OSD-44 and OSD-121). Most studies are categorized based on organism and tissue type, although plant studies are distinguished based solely on organism as shown here. Each study is further categorized between ‘omics and phenotypic-physiological datasets. GLDS includes all ‘omics data and LSDS contains all phenotypic-physiological data derived from the respective study. — NASA
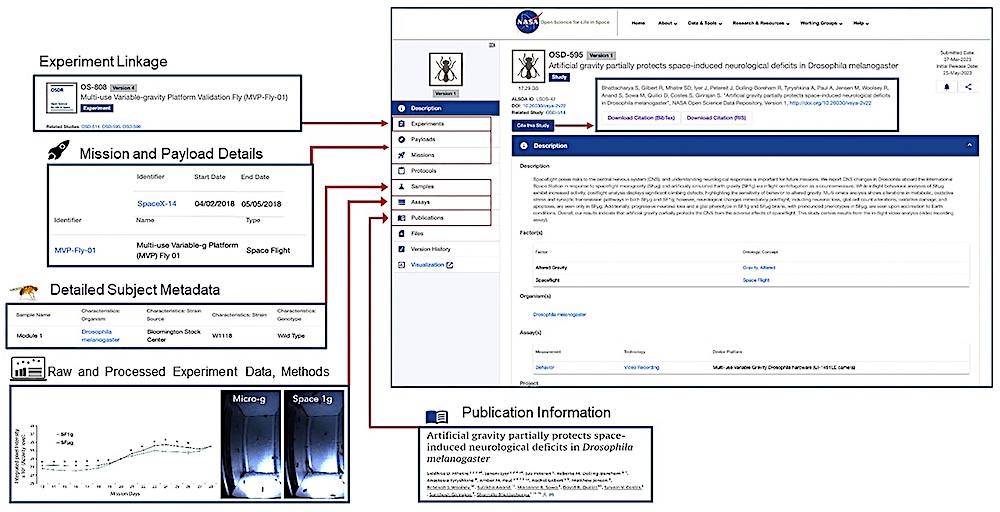
An OSD study page. Screenshot of study OSD-595 showing the links to the respective experiments (OS-808), mission (SpaceX-14) and payload (MVP-Fly-01) details, sample metadata, GLDS and LSDS assay metadata and primary publications (left) and study citation (inset). — NASA
- NASA open science data repository: open science for life in space, Nucleic Acids Research (open access)
- NASA Open Science Data Repository
- NASA Life Sciences Portal (NLSP)
- Celebrating 30 Years Of Access To NASA Space Life Sciences Data
Astrobiology,