Mapping the Edges of Mass Spectral Prediction: Evaluation of Machine Learning EIMS Prediction for Xeno Amino Acids
Mass spectrometry is one of the most effective analytical methods for unknown compound identification. By comparing observed m/z spectra with a database of experimentally determined spectra, this process identifies compound(s) in any given sample. Unknown sample identification is thus limited to whatever has been experimentally determined.
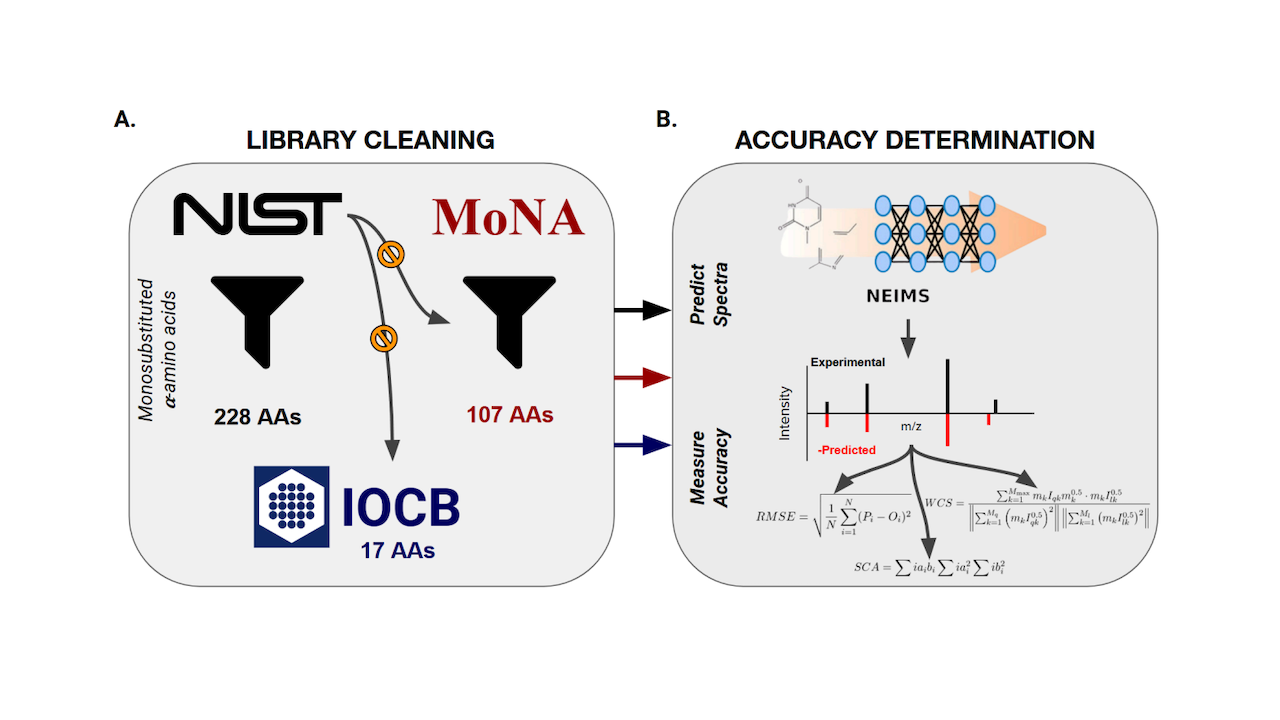
To address the reliance on experimentally determined signatures, multiple state-of-the-art MS spectra prediction algorithms have been developed within the past half decade. Here we evaluate the accuracy of the NEIMS spectral prediction algorithm.
We focus our analyses on monosubstituted α-amino acids given their significance as important targets for astrobiology, synthetic biology, and diverse biomedical applications. Our general intent is to inform those using generated spectra for detection of unknown biomolecules.
We find predicted spectra are inaccurate for amino acids beyond the algorithms training data. Interestingly, these inaccuracies are not explained by physicochemical differences or the derivatization state of the amino acids measured.
We thus highlight the need to improve both current machine learning based approaches and further optimization of ab initio spectral prediction algorithms so as to expand databases for structures beyond what is currently experimentally possible, even including theoretical molecules.
Mapping the Edges of Mass Spectral Prediction: Evaluation of Machine Learning EIMS Prediction for Xeno Amino Acids, chemrxiv.org
Astrobiology,