Henneguya salminicola Lacks A Mitochondrial Genome And Does Not Require Oxygen
Mitochondrial respiration is an ancient characteristic of eukaryotes. However, it was lost independently in multiple eukaryotic lineages as part of adaptations to an anaerobic lifestyle.
We show that a similar adaptation occurred in a member of the Myxozoa, a large group of microscopic parasitic animals that are closely related to jellyfish and hydroids. Using deep sequencing approaches supported by microscopic observations, we present evidence that an animal has lost its mitochondrial genome.
The myxozoan cells retain structures deemed mitochondrion-related organelles, but have lost genes related to aerobic respiration and mitochondrial genome replication. Our discovery shows that aerobic respiration, one of the most important metabolic pathways, is not ubiquitous among animals.
Abstract
Although aerobic respiration is a hallmark of eukaryotes, a few unicellular lineages, growing in hypoxic environments, have secondarily lost this ability. In the absence of oxygen, the mitochondria of these organisms have lost all or parts of their genomes and evolved into mitochondria-related organelles (MROs).
There has been debate regarding the presence of MROs in animals. Using deep sequencing approaches, we discovered that a member of the Cnidaria, the myxozoan Henneguya salminicola, has no mitochondrial genome, and thus has lost the ability to perform aerobic cellular respiration.
This indicates that these core eukaryotic features are not ubiquitous among animals. Our analyses suggest that H. salminicola lost not only its mitochondrial genome but also nearly all nuclear genes involved in transcription and replication of the mitochondrial genome.
In contrast, we identified many genes that encode proteins involved in other mitochondrial pathways and determined that genes involved in aerobic respiration or mitochondrial DNA replication were either absent or present only as pseudogenes. As a control, we used the same sequencing and annotation methods to show that a closely related myxozoan, Myxobolus squamalis, has a mitochondrial genome. The molecular results are supported by fluorescence micrographs, which show the presence of mitochondrial DNA in M. squamalis, but not in H. salminicola.
Our discovery confirms that adaptation to an anaerobic environment is not unique to single-celled eukaryotes, but has also evolved in a multicellular, parasitic animal. Hence, H. salminicola provides an opportunity for understanding the evolutionary transition from an aerobic to an exclusive anaerobic metabolism.
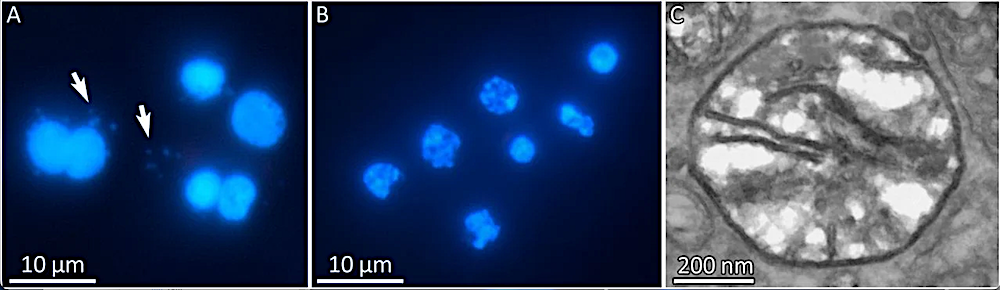
Microscopic evidence for the absence of mitochondria in H. salminicola. (A and B) DAPI staining of normal 7-cell presporogonic developmental stages of two myxozoan parasites of salmonid fish. (A) M. squamalis, showing large nuclei with many smaller mitochondrial nucleosomes (arrowed). (B) H. salminicola, showing large nuclei but surprisingly no mitochondrial nucleosomes. (C) TEM image of H. salminicola mitochondrion-related organelle with few cristae. Uncropped images are available in the Figshare repository. — PNAS via PubMed
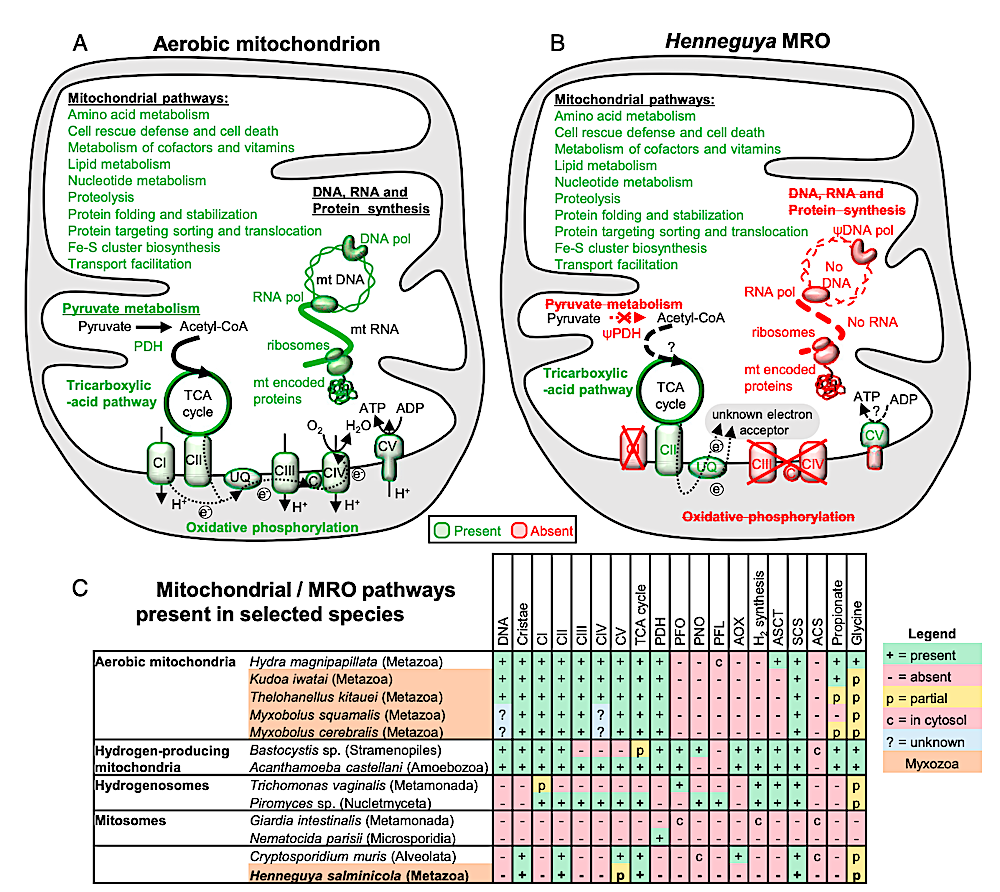
Comparison between the pathways present in (A) a typical aerobic mitochondrion and (B) the H. salminicola MRO. (C) Mitochondrial/MRO pathways present in selected species (see refs. 1 and 2). The presence and absence of organellar genomes are indicated. ACS, acetyl-CoA synthetase; acetate AOX, alternative oxidase; ASCT, acetate succinyl-CoA transferase; DNA pol, mtDNA polymerase; RNA pol, mtDNA-dependent RNA polymerase; CI-CV, respiratory complexes I-V; C, cytochrome c; PDH, pyruvate dehydrogenase; PFL, pyruvate formate lyase; PFO, pyruvate ferredoxin oxidoreductase; PNO, pyruvate NADPH oxidoreductase; SCS, succinyl-CoA synthetase; TCA cycle, tricarboxylic acid cycle; UQ, ubiquinone; e− , electrons; H+ , protons; ψ indicates the presence of a pseudogene in the nuclear genome. — PNAS via PubMed
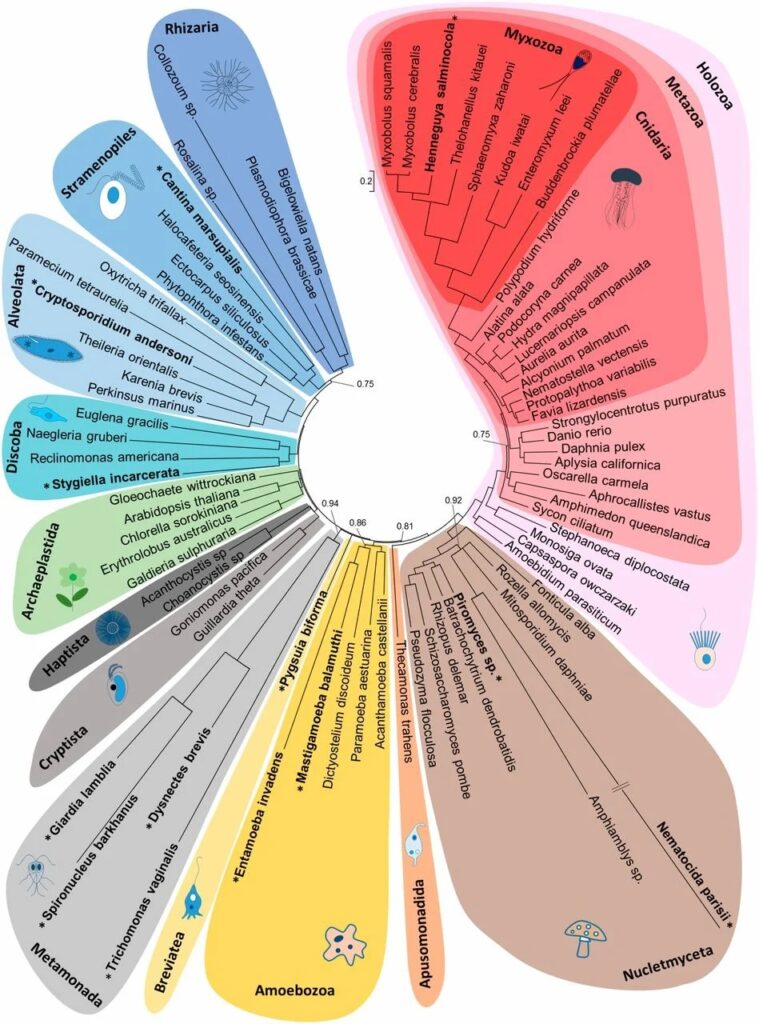
Eukaryote phylogenetic relationships inferred from a supermatrix of 9490 amino acid positions for 78 species. Bayesian majority-rule consensus tree reconstructed using the CAT + Γ model from two independent Markov-chain Monte Carlo chains. Branches with low node support (posterior probabilities PP < 0.7) were collapsed. Most nodes were highly supported (PP > 0.98), and PP are only indicated for nodes with PP < 0.98. The eukaryote classification is based on Adl et al. (47). Species known to have lost their mt genome are indicated in bold with an asterisk. Myxozoan species form a well-supported group (PP = 1.0) and our reconstructions agree with previous studies (14), which show monophyly of the fresh-water/oligochaete host lineage (10).— PNAS via PubMed
- A cnidarian parasite of salmon (Myxozoa: Henneguya) lacks a mitochondrial genome, PNAS via PubMed
- Unique Non-oxygen Breathing Animal Discovered, earlier post
astrobiology