Previously Unknown Life Forms Discovered Within Human Microbiomes
Editor’s note: A team of researchers at Stanford University discovered objects they named “obelisks” in the human mouth and gut. The obelisks are microscopic, circular bits of genetic material that contain one or two genes. They self-organize into a rod-like shape, which is how they got their name. Obelisks are surprisingly common in the human microbiome – the community of viruses, bacteria, fungi, and their genes that live in our bodies. The function of obelisks is a mystery, and they may represent an entirely new class of life. However, they could hold the key to understanding life itself, and as research continues, we may learn more about how they impact our health and their origins. The fact that we can make surprising discoveries of potentially new types of Earth life – or perhaps just especially unusual ones – inside our own bodies – after decades of study – speaks to the unexpected diversity of life on our own world, and it certainly serves as a reminder of what vast varieties of life may await us on other worlds.
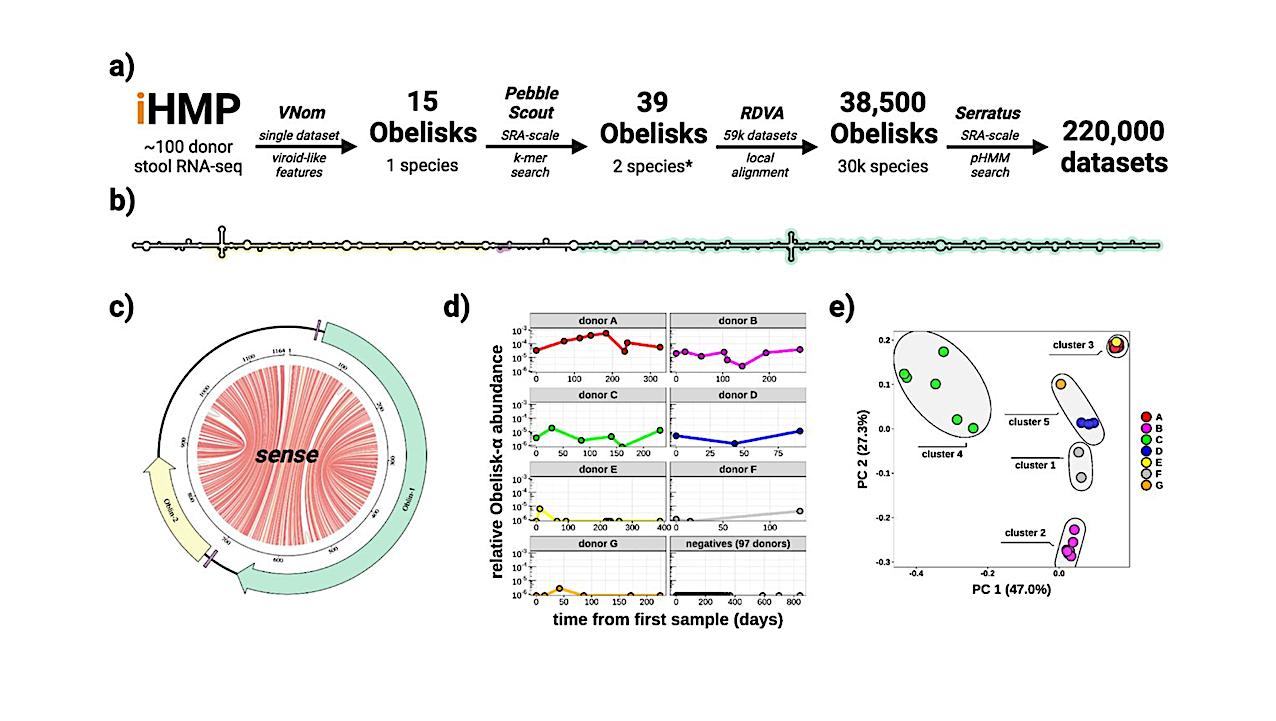
Here, we describe the “Obelisks,” a previously unrecognised class of viroid-like elements that we first identified in human gut metatranscriptomic data. “Obelisks” share several properties: (i) apparently circular RNA ∼1kb genome assemblies, (ii) predicted rod-like secondary structures encompassing the entire genome, and (iii) open reading frames coding for a novel protein superfamily, which we call the “Oblins”.
We find that Obelisks form their own distinct phylogenetic group with no detectable sequence or structural similarity to known biological agents.
Further, Obelisks are prevalent in tested human microbiome metatranscriptomes with representatives detected in ∼7% of analysed stool metatranscriptomes (29/440) and in ∼50% of analysed oral metatranscriptomes (17/32).
Obelisk compositions appear to differ between the anatomic sites and are capable of persisting in individuals, with continued presence over >300 days observed in one case. Large scale searches identified 29,959 Obelisks (clustered at 90% nucleotide identity), with examples from all seven continents and in diverse ecological niches.
From this search, a subset of Obelisks are identified to code for Obelisk-specific variants of the hammerhead type-III self-cleaving ribozyme. Lastly, we identified one case of a bacterial species (Streptococcus sanguinis) in which a subset of defined laboratory strains harboured a specific Obelisk RNA population.
As such, Obelisks comprise a class of diverse RNAs that have colonised, and gone unnoticed in, human, and global microbiomes.
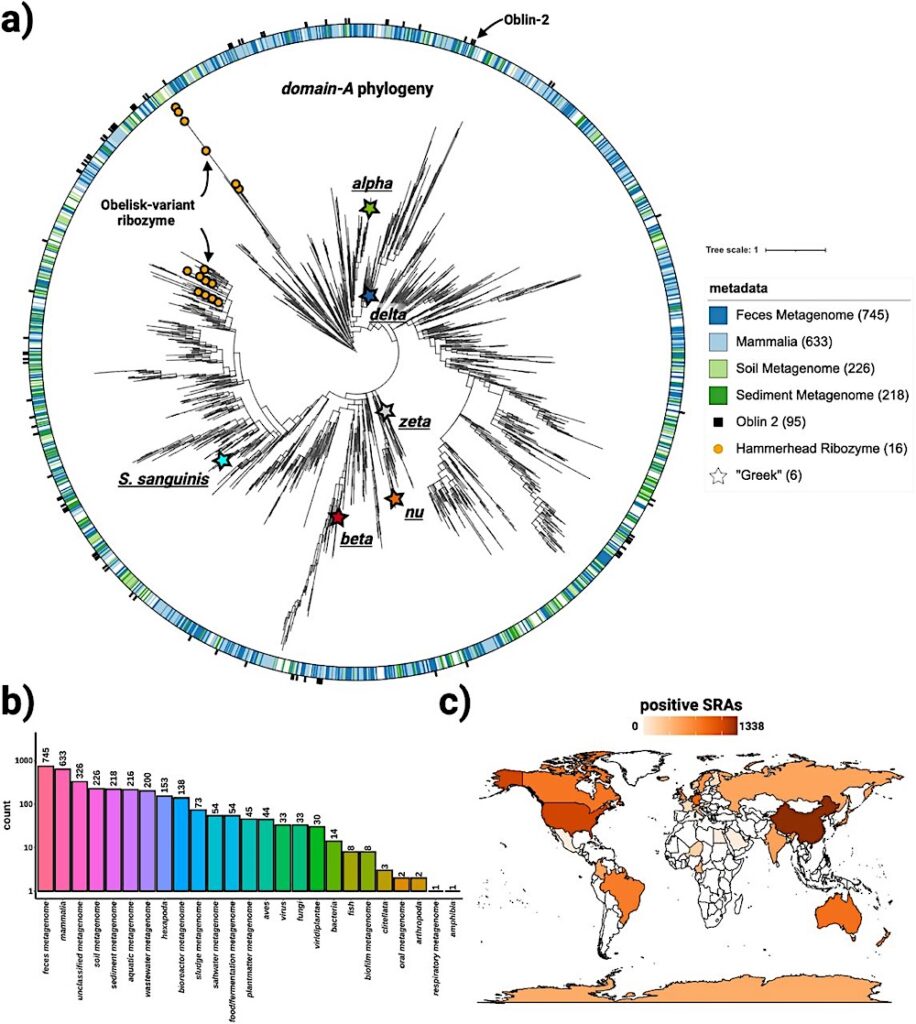
Obelisks form their own globally distributed phylogenetic group a) a maximum likelihood, midpoint-rooted, phylogenetic tree (see methods) constructed from a non-redundant set of 3265 Serratus and RDVA domain-A sequences, with RDVA genomes positive for Obelisk-variant self cleaving Hammerhead Type III ribozymes illustrated as orange circles on leaves, and the top four known classes of SRA “host” metadata depicted as the colour band (see legend), and with per-RDVA-genome co-occurrence of Oblin-2 (based on blastp hits against the Oblin-2 consensus) illustrated as the outer ring (black studs). Leaves that correspond to domain-A sequences from Figure 4 are illustrated with stars. b) Counts of non de-replicated SRA datasets used to construct a) sorted by their “host” metadata; we note that “host” metadata likely fails to account other organisms’ genetic material that was sequences alongside the “host” (e.g. signals from these hosts’ microbiomes maybe be detected in tandem). c) Counts of non de-replicated SRA datasets used to construct a) arranged by sample geolocation (where known) illustrated on a world map (darker orange = more SRA datasets contributed to a)). We note that SRA counts are not expected to correlate with true geo-/ecological prevalence, but are still indicative of global presence. — Cell via PubMed
Viroid-like colonists of human microbiomes, Cell via biorxiv.org
Astrobiology