Nitrogenase Structural Evolution Across Earth’s History
Life on Earth is more than 3.5 billion years old—nearly as old as the age of the planet. Over this vast expanse of time, life and its biomolecules adapted to and triggered profound changes to the Earth’s environment
Certain critical enzymes evolved early in the history of life and have persisted through planetary extremes. While sequence data is widely used to trace evolutionary trajectories, enzyme structure remains an underexplored resource for understanding how proteins evolve over long timescales.
Here, we implement an integrated approach to study nitrogenase, an ancient, globally critical enzyme essential for nitrogen fixation. Despite the ecological diversity of its host microbes, nitrogenase has strict functional limitations, including extreme oxygen sensitivity, energy requirements and substrate availability.
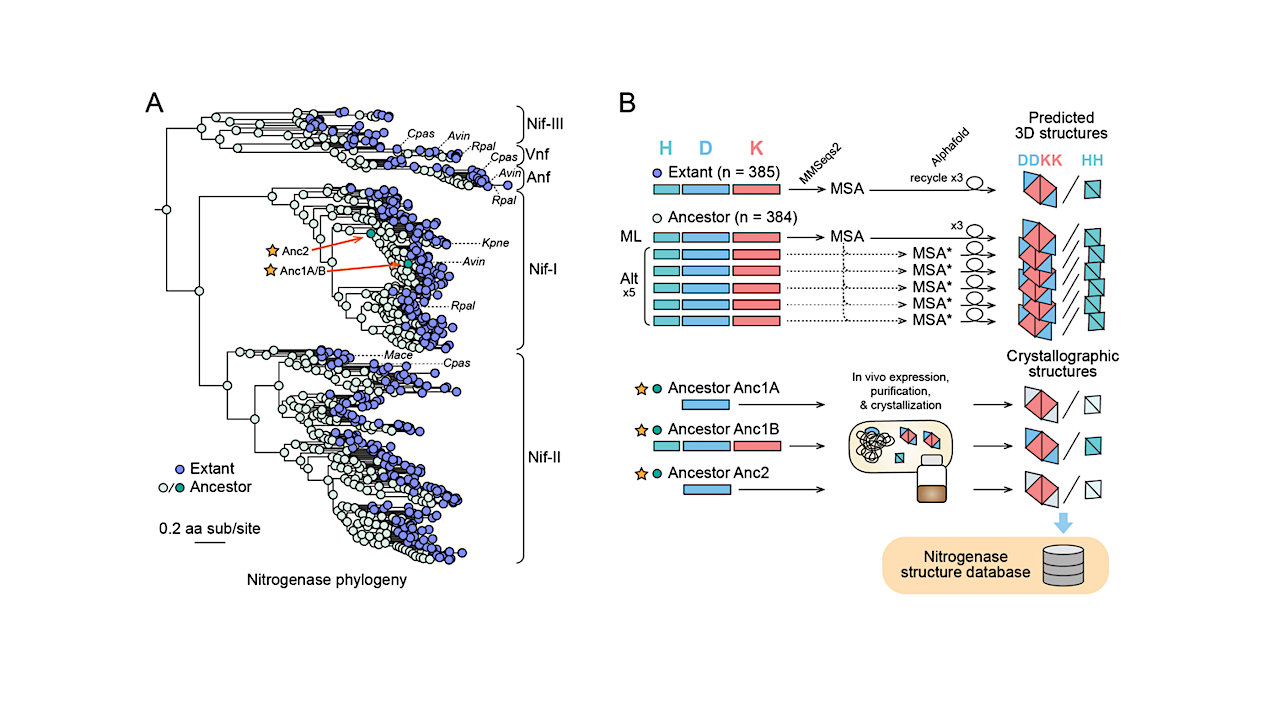
By combining phylogenetics, ancestral sequence reconstruction, protein crystallography and deep-learning based structural prediction, we resurrected three billion years of nitrogenase structural history.
We present the first effort to predict all extant and ancestral structures along the evolutionary tree of an enzyme and present a total of ∼5000 structures. Our approach lays the foundation for reconstructing key structural constraints that influence protein evolution and studying ancient enzyme evolution in the light of phylogenetic and environmental change.
Nitrogenase structural evolution across Earth’s history, biorxiv.org (open access)
Astrobiology,