New Insight Into Chloroplast Evolution
One of the most momentous events in the history of life involved endosymbiosis — a process by which one organism engulfed another and, instead of ingesting it, incorporated its DNA and functions into itself. Scientific consensus is that this happened twice over the course of evolution, resulting in the energy-generating organelles known as mitochondria and, much later, their photosynthetic counterparts, the plastids.
A new study published in the journal Nature Communications explores the origin of chloroplasts, the plastids that allow plants to extract carbon from the atmosphere to build their own structures and tissues. By focusing on an energy-transport molecule common to plastids, the researchers found evidence suggesting that the primary role of primitive chloroplasts may have been to produce chemical energy for the cell and only later shifted so that most or all of the energy they generated was used for carbon assimilation.
Chloroplasts are believed to have evolved from photosynthetic cyanobacteria, but it isn’t clear what functions the cyanobacteria originally performed for the cells that engulfed them, said University of Illinois Urbana-Champaign chemistry professor Angad Mehta, who led the new research.
“We asked the question: What chemical role did the primitive symbiont that led to chloroplasts perform for the host cell?” he said. “Was it carbon assimilation or ATP synthesis or both?”
Various lines of evidence suggest that the plastids in red algae and another group of photosynthesizing organisms known as glaucophytes resemble more ancient stages of evolution than the chloroplasts of land plants. But current bioinformatics methods can take the field only so far, Mehta said.
A key to the functional evolution of mitochondria and plastids lies in their energy-generating capacities, he said. Both produce ATP, an energy-packed molecule that drives most of the chemical interactions in living cells. And both mitochondria and plastids make use of ADP/ATP carrier translocases, which reside in the membranes of the organelles and swap ATP with its energy-depleted precursor, ADP.
Mehta and his colleagues focused on differences in the activity of the translocases in the plastids of land plants, red algae and glaucophytes to determine whether these differences could offer insight into chloroplast evolution.
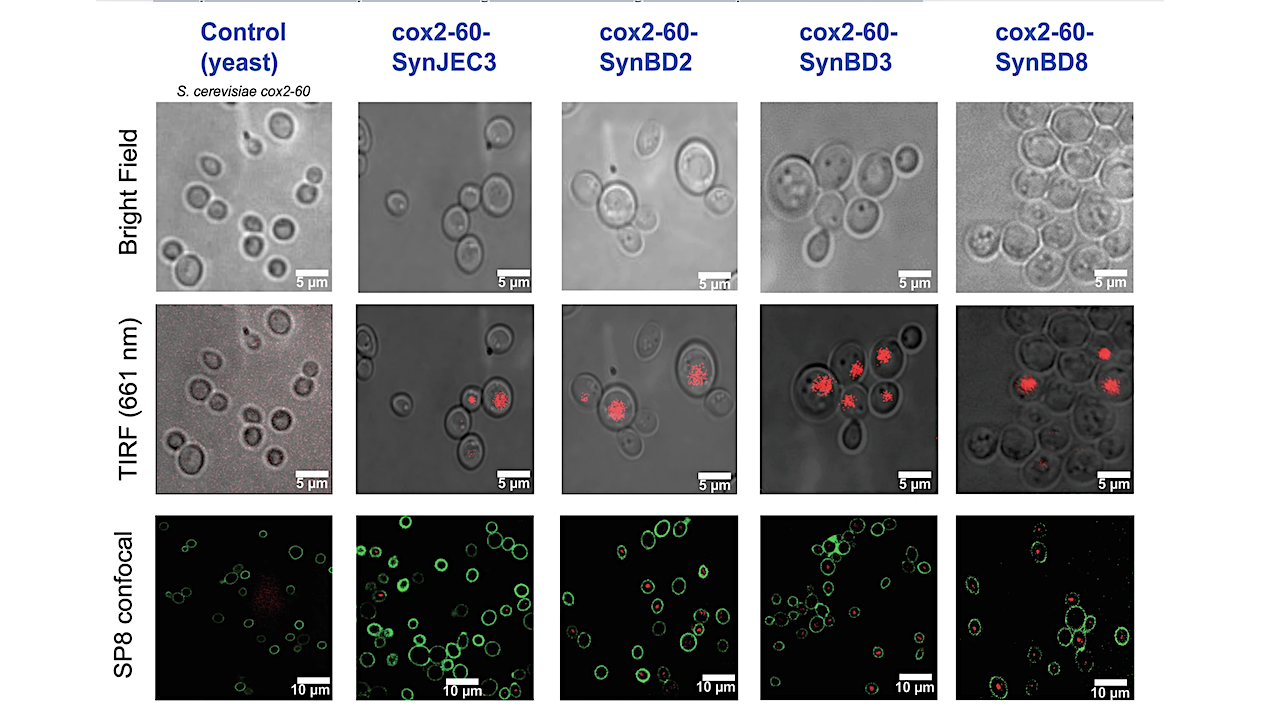
In a series of experiments, the researchers engineered cyanobacteria to express one of the three types of translocases. Then they induced artificial endosymbiosis between the engineered cyanobacteria and budding yeast cells. By controlling the laboratory conditions in which these cells lived, the researchers forced the yeast to rely entirely on the cyanobacterial endosymbionts for their energy needs. Mehta’s lab first developed the technique for artificially forcing yeast to internalize cyanobacterial endosymbionts in a study published in 2022.
The experiments revealed striking differences between the activity of the translocases.
“Most notably, we saw that the endosymbionts expressing translocases from the plastids of red algae and glaucophytes were able to export ATP to support endosymbiosis, whereas those from chloroplasts actually imported ATP and were unable to support the energy needs of the endosymbiotic cells,” Mehta said. The land plant chloroplast translocases were importing ATP and expelling ADP.
Because the plastids of red algae and glaucophytes appear to resemble a more ancient form of the photosynthetic organelles, the new findings suggest that chloroplasts once shared their primary function of providing energy to the larger cell. At some point in their evolutionary history, however, the chloroplasts of land plants appear to have shifted to use the ATP they produced via photosynthesis to drive their own carbon-assimilation tasks. It appears that chloroplasts even siphon off some of the ATP generated by mitochondria, Mehta said.
While the new findings do not definitively prove that this is how chloroplasts evolved, it does offer evidence to support this view, Mehta said.
“The proposal is that the initial interaction between the endosymbiont and cell was based on ATP production and ATP supply,” he said. “Now, you can imagine a scenario in which, as these organisms go on to become land plants, they grow in oxygen-rich conditions. This allows the mitochondria to become specialized in ATP synthesis and chloroplasts to focus and become an engine that drives carbon assimilation.”
The Gordon and Betty Moore Foundation and the National Institute of General Medical Sciences at the National Institutes of Health supported this research. Mehta also is an affiliate of the Carl R. Woese Institute for Genomic Biology at the U. of I.
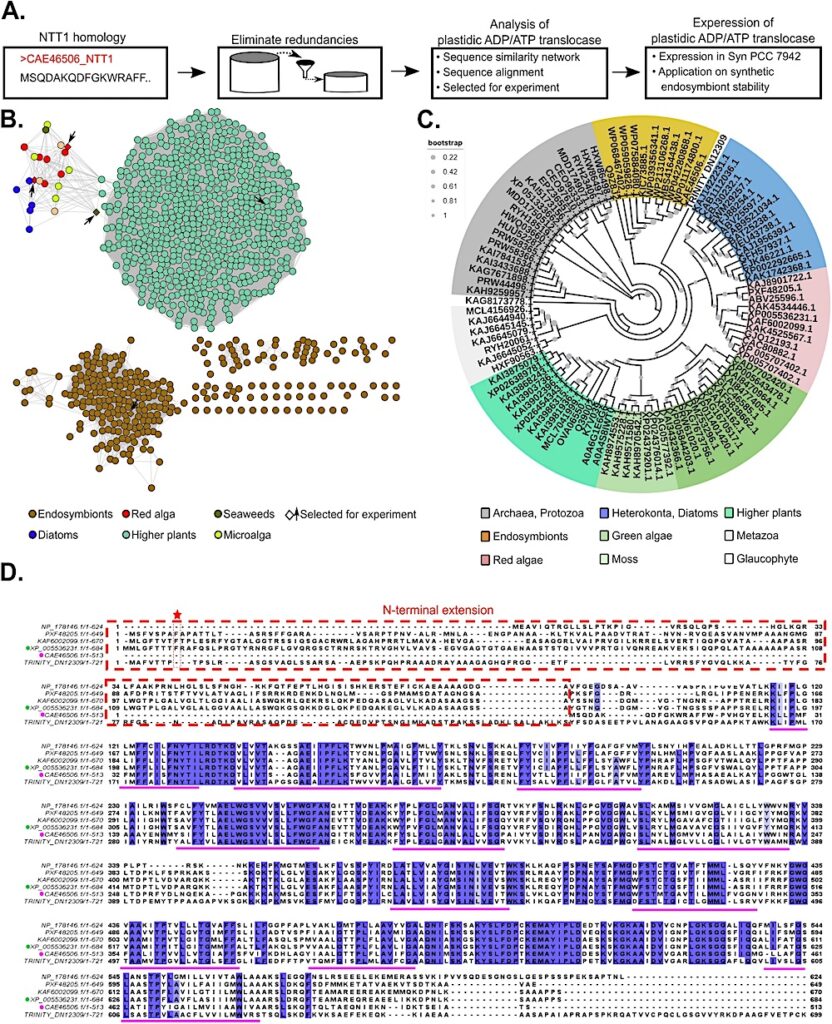
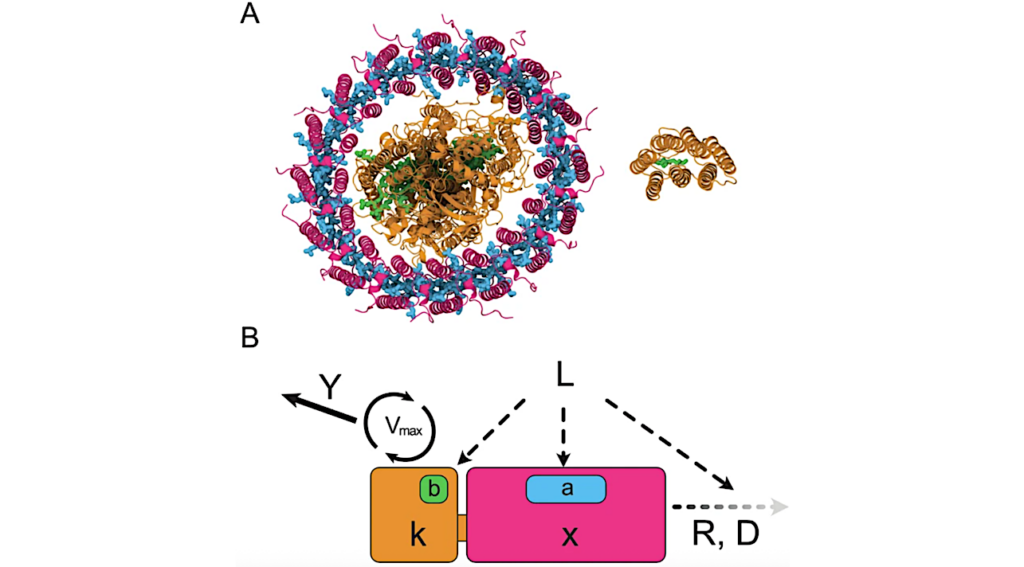
A The general workflow for identification and characterization of plastidic ADP/ATP carrier translocases. B Sequence similarity network (SSN) for ADP/ATP carrier translocase proteins found across the photosynthetic eukaryotes; the SSN is analyzed using an alignment score 120. The NTT proteins from Endosymbionts (Brown), and the ADP/ATP carrier translocase proteins from red algae (Red), diatoms (blue), microalgae/green algae (light green), higher plants (teal), the selected sequence from the SSN were presented rhombus border. C A smaller data set phylogenetic analysis of putative primary plastid ADP/ATP carrier translocase family proteins from archea to higher plant (see Results and Methods section). D Amino acid sequence alignment of selected NTT homologous and orthologs of putative plastidic ADP/ATP carrier translocase family protein from red algae and higher plant. The multiple sequence alignment was performed using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) and data was presented using jalview software. Colored boxes indicate conserved amino acid residues among NTT and putative plastidic ADP/ATP carrier translocase family proteins from red algae and higher plant (100%: fore black, back blue; 80%–60%: fore black, back light blue). The pink dot represent amino acid query sequence: CAE46506.1; the Green dot amino acid sequence of plastidic putative ADP/ATP carrier protein (Cyanidioschyzon merolae strain 10D) NCBI id: XP_005536231.1 and its corresponding PDB id: M1V528; the dotted red rectangular box represented the N-terminal extension of putative plastidic ADP/ATP carrier translocase family proteins from red algae, glaucophyte and higher plant; the red star represented plastidic signal peptide conserved phenylalanine (F) essential organelle targeting; the pink underline represented the predicted twelve transmembrane domain for plastidic putative ADP/ATP carrier protein (C. merolae strain 10D) PDB id: M1V528. Source data are provided as a Source Data file. — Nature
The paper “Photosynthetic directed endosymbiosis to investigate the role of bioenergetics in chloroplast function and evolution”. Nature Communications is available online open access DOI: 10.1038/s41467-024-54051-1
Astrobiology