Mirror Bacteria Report: Call For A Global Discussion About Possible Risks
Editor’s note: As we study how Earth life is put together it is helpful to understand alternate ways that chemistry could combine to form living systems. One way is to use the mirror image of all the molecular components of Earth life. There are concerns that such an artificial tree of life could endanger our ecosystems and all of the life forms that comprise them. Hence the concerns voiced in this report. One thing to keep in mind: while our world has evolved with its own biochemistry the life chemistries of other worlds may not necessarily be something that we want to interact with – or on the other hand – contaminiate.
A group of researchers has published new findings in Science on potential risks from the development of mirror bacteria — synthetic organisms in which all molecules have reversed chirality (i.e. are ‘mirrored’).
Scientists had begun early work toward creating mirror bacteria, and while the capability is at least a decade away, recent years have seen significant progress. The new paper finds that, if created, these organisms may pose significant dangers to human, animal, plant, and environmental health. The authors call for a broad conversation among scientists, policymakers, and a wide range of other stakeholders to chart a path towards better understanding and mitigation of potential risks from mirror bacteria.
The 38 authors working in nine countries include leading experts in immunology, plant pathology, ecology, evolutionary biology, biosecurity, and planetary sciences. The publication in Science is accompanied by a detailed 300-page technical report.
- “Technical Report on Mirror Bacteria: Feasibility and Risks“, Standford University
While any threat is not imminent, the Science paper finds that mirror bacteria may pose serious risks. Immune defenses in humans, animals, and plants rely on recognizing specific molecular shapes found in invading bacteria. If these shapes were reflected — as they would be in mirror bacteria — recognition would be impaired and many basic immune defenses could fail, potentially leaving organisms vulnerable to infection.
The analysis also suggests that mirror bacteria in the environment may be able to evade natural predators like phages and protists, which rely heavily on chirally-mediated interactions to kill bacteria and limit their populations. Transport via animals and humans could enable spread between diverse ecosystems. Persistent and widespread environmental populations of mirror bacteria would expose humans, animals and plants to an ongoing risk of infection — a serious threat to humans and to global ecosystems.
The authors call for further scrutiny of their findings and conclude that, unless compelling evidence emerges that these organisms would not pose extraordinary dangers, mirror bacteria should not be created. Notably, the group includes several authors who previously held the creation of mirror bacteria as a long-term aspirational goal.
This paper marks a starting point for a broader discussion about the risks from mirror bacteria, including participation from the global scientific community, policymakers, research funders, and other stakeholders. Several of the authors on the paper are involved in planning a series of events throughout 2025, including events planned at the Institut Pasteur in France, the University of Manchester in the U.K. and the National University of Singapore, to scrutinize the findings of the paper and discuss steps that can be taken to prevent risks from mirror bacteria.
Confronting risks of mirror life, Science
“Technical Report on Mirror Bacteria: Feasibility and Risks“, Stanford University
“This report describes the technical feasibility of creating mirror bacteria and the potentially serious and wide-ranging risks that they could pose to humans, other animals, plants, and the environment. It accompanies the Science Policy Forum article titled “Confronting risks of mirror life”, published December 12, 2024.
In a mirror bacterium, all of the chiral molecules of existing bacteria—proteins, nucleic acids, and metabolites—are replaced by their mirror images. Mirror bacteria could not evolve from existing life, but their creation will become increasingly feasible as science advances. Interactions between organisms often depend on chirality, and so interactions between natural organisms and mirror bacteria would be profoundly different from those between natural organisms. Most importantly, immune defenses and predation typically rely on interactions between chiral molecules that could often fail to detect or kill mirror bacteria due to their reversed chirality. It therefore appears plausible, even likely, that sufficiently robust mirror bacteria could spread through the environment unchecked by natural biological controls and act as dangerous opportunistic pathogens in an unprecedentedly wide range of other multicellular organisms, including humans.
This report draws on expertise from synthetic biology, immunology, ecology, and related fields to provide the first comprehensive assessment of the risks from mirror bacteria. It consists of eight chapters and starts with a general introduction, followed by an examination of the initial creation of mirror bacteria, their further engineering, as well as biosecurity and biosafety implications. The remaining five chapters cover risks to human health, medical countermeasures, risks to other animals, risks to plants, and the potential ecological consequences of their introduction into the environment.”
IMAGE
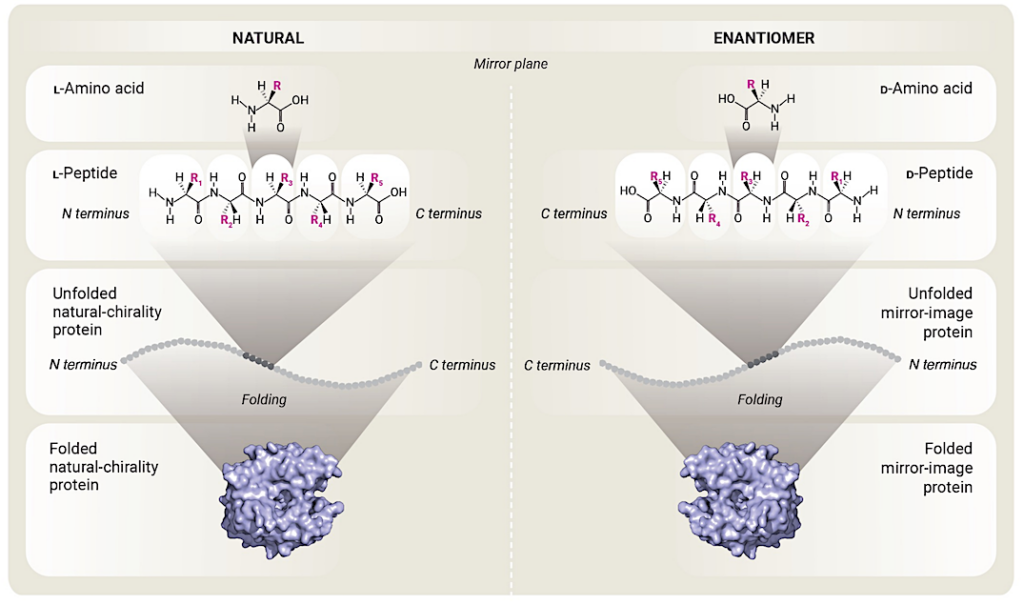
Mirror-image amino acids compose mirror-image peptides that fold into mirror-image proteins All natural amino acids except glycine are chiral, and only the ʟ-enantiomers are used in natural proteins. Substituting the ʟ-amino acids for their mirror image ᴅ-amino acid enantiomers would result in a mirror-image polypeptide chain. The interactions between the side chains for amino acids in the ᴅ-peptide are the same as the ʟ-peptide, but with the opposite orientation. Thus, the mirror-image peptide chain folds into a protein that is the mirror image of its natural chirality counterpart. (Structure image for example protein, carbonic anhydrase, produced by Pymol 3.0.0 from Protein Data Bank accession number 1CA2.) — Stanford University
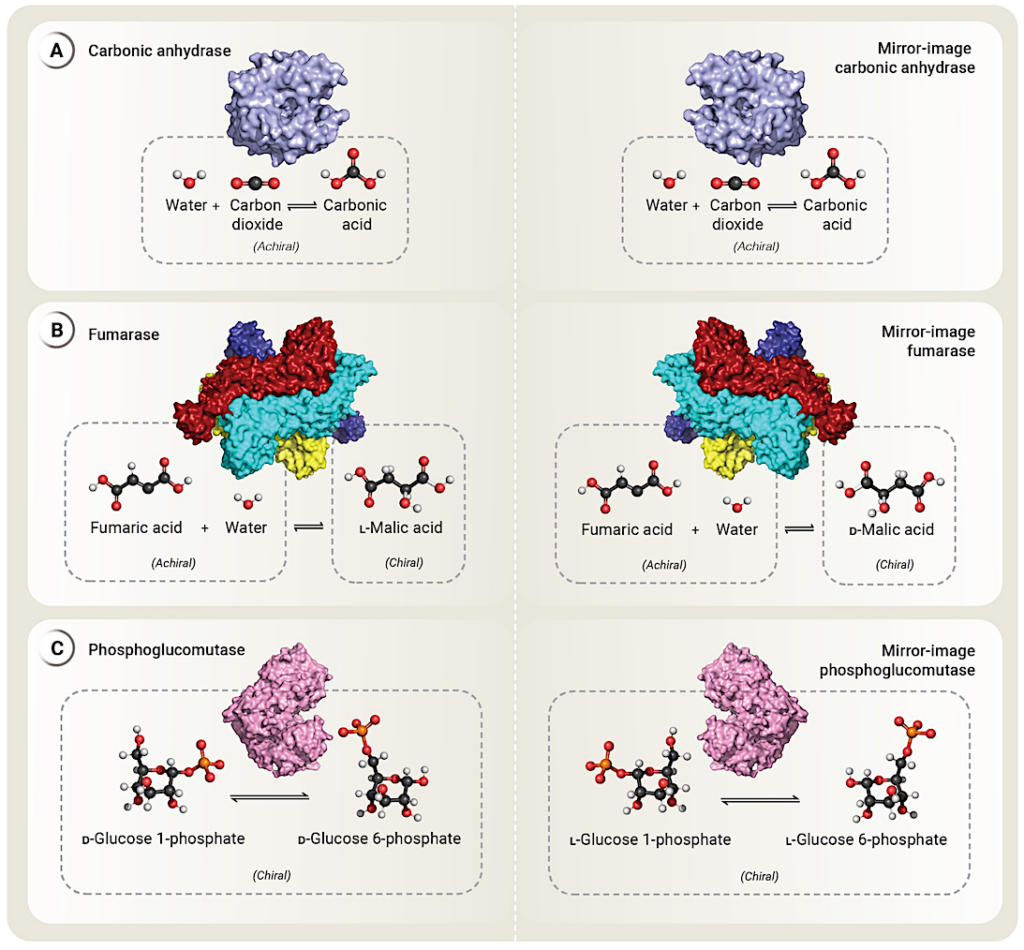
Mirror-image enzymes catalyze mirror-image reactions Enzyme catalysis requires precise (and thus stereospecific) binding of substrate molecules to the active site. A. If all substrates and products of an enzyme are achiral, such as for carbonic anhydrase, the natural-chirality and mirror-image enzymes would behave identically. B. When the products of an enzymatic reaction are chiral, the natural-chirality enzyme exclusively produces one enantiomer and its mirror-image enzyme would produce the other. For example, both fumarase and mirror-image fumarase would produce malic acid from fumaric acid and water, but natural-chirality fumarase exclusively produces ʟ-malic acid (the enantiomer seen in nature), while mirror-image fumarase would exclusively produce ᴅ-malic acid. C. When the substrate of an enzyme is chiral, catalysis is typically specific to that enantiomer. For example, natural-chirality phosphoglucomutase can convert ᴅ-glucose 1-phosphate to ᴅ-glucose 6-phosphate. In contrast, mirror-image phosphoglucomutase would convert ʟ-glucose 1-phosphate to ʟ-glucose 6-phosphate. (Structure images for carbonic anhydrase, fumarase, and phosphoglucomutase produced by Pymol 3.0.0 from Protein Data Bank accession numbers 1CA2, 3E04, and 3PMG respectively.) — Stanford University
IMAGE
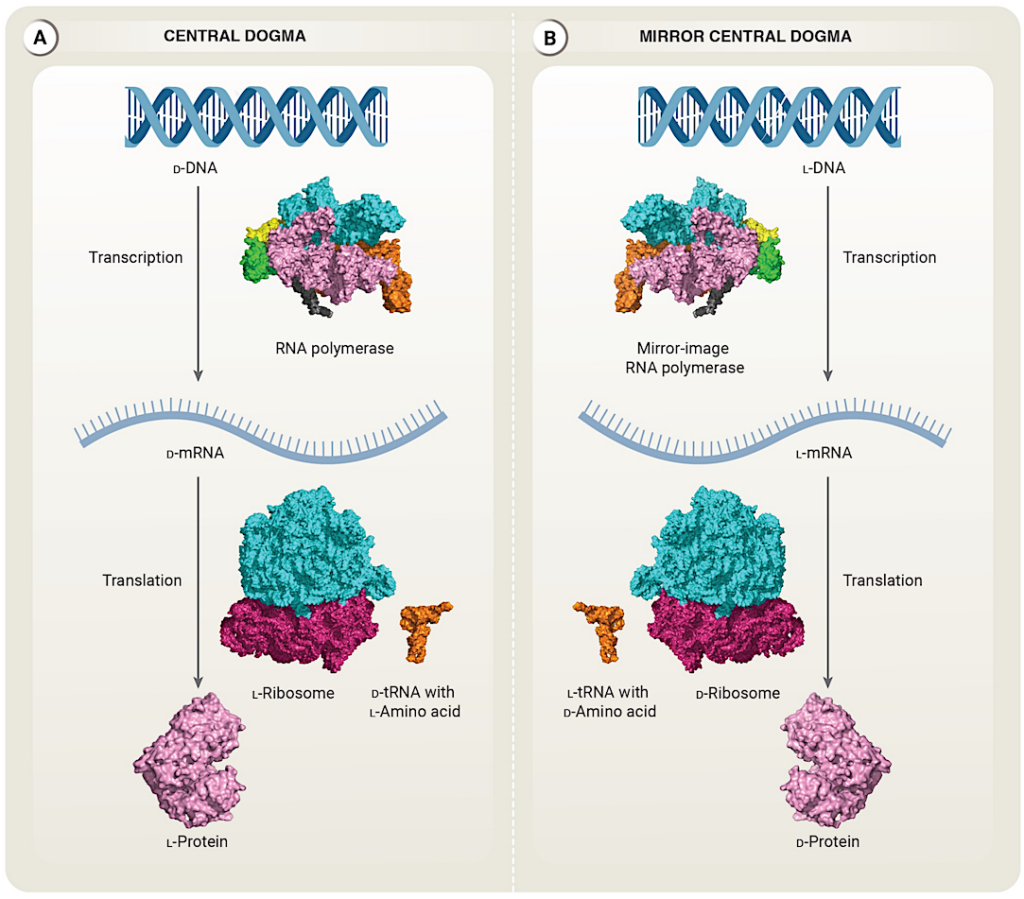
A mirror image of the central dogma A. The central dogma of molecular biology. Genetic information in the form of ᴅ-DNA is transcribed by natural-chirality RNA to ᴅ-messenger RNA (ᴅ-mRNA), which is in turn translated into natural-chirality polypeptides and proteins. All of these stages exclusively use one of the two possible chiralities: natural chirality RNA polymerase transcribes ᴅ-DNA into ᴅ-mRNA, which the natural-chirality ribosome and transfer RNA (tRNA) carrying ʟ-amino acids then translate into ʟ-proteins. B. The central dogma could be chirally inverted: ʟ-DNA would be transcribed by a mirror-image RNA polymerase into ʟ-mRNA, which would be translated into mirror-image ᴅ-proteins. (Structure images for RNA polymerase, the ribosome, tRNA, and example protein (phosphoglucomutase) produced by Pymol 3.0.0 from Protein Data Bank accession numbers 4YG2, 5AFI, 6UGG, and 3PMG respectively.) — Stanford University
Astrobiology,