Tricorder Tech: Multi-pass, Single-molecule Nanopore Reading of Long Protein Strands
Editor’s note: Oxford Nanopore technology has been used multiple times on the International Space Station (see Genomics, Proteomics, Bioinformatics) to try and help elucidate changes in genomic function during exposure to conditions of spaceflight. This technology has also been used to track down the source of diseases in remote areas, characterize extremophiles in Antarctica, and study sources of food contamination. As we prepare for Away Team traverses on other worlds advanced versions of this sort of sequencing technology will be essential components of a crew’s toolset.
The ability to sequence single protein molecules in their native, full-length form would enable a more comprehensive understanding of proteomic diversity.
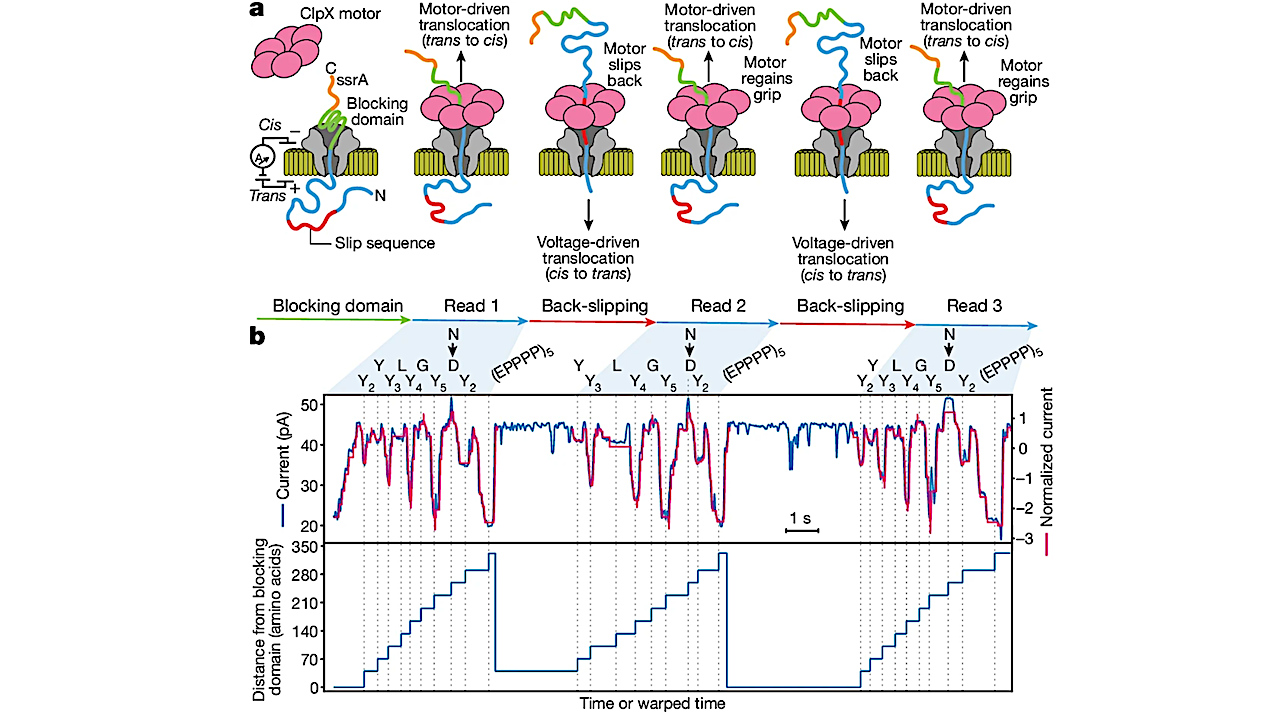
Current technologies, however, are limited in achieving this goal. Here, we establish a method for the long-range, single-molecule reading of intact protein strands on a commercial nanopore sensor array. By using the ClpX unfoldase to ratchet proteins through a CsgG nanopore, we provide single-molecule evidence that ClpX translocates substrates in two-residue steps.
This mechanism achieves sensitivity to single amino acids on synthetic protein strands hundreds of amino acids in length, enabling the sequencing of combinations of single-amino-acid substitutions and the mapping of post-translational modifications, such as phosphorylation. To enhance classification accuracy further, we demonstrate the ability to reread individual protein molecules multiple times, and we explore the potential for highly accurate protein barcode sequencing.

Furthermore, we develop a biophysical model that can simulate raw nanopore signals a priori on the basis of residue volume and charge, enhancing the interpretation of raw signal data.
Finally, we apply these methods to examine full-length, folded protein domains for complete end-to-end analysis. These results provide proof of concept for a platform that has the potential to identify and characterize full-length proteoforms at single-molecule resolution.
Multi-pass, single-molecule nanopore reading of long protein strands, Nature via PubMed
PMID: 39261738 PMCID: PMC11410661 DOI: 10.1038/s41586-024-07935-7 (open access)
Astrobiology, genomics,