Ocean Planet Exploration Droids: Autonomous eDNA Collection Using An Uncrewed Surface Vessel
The collection of environmental DNA (eDNA) samples is often laborious, costly, and logistically difficult to accomplish at high frequency in remote locations and over large geographic areas.
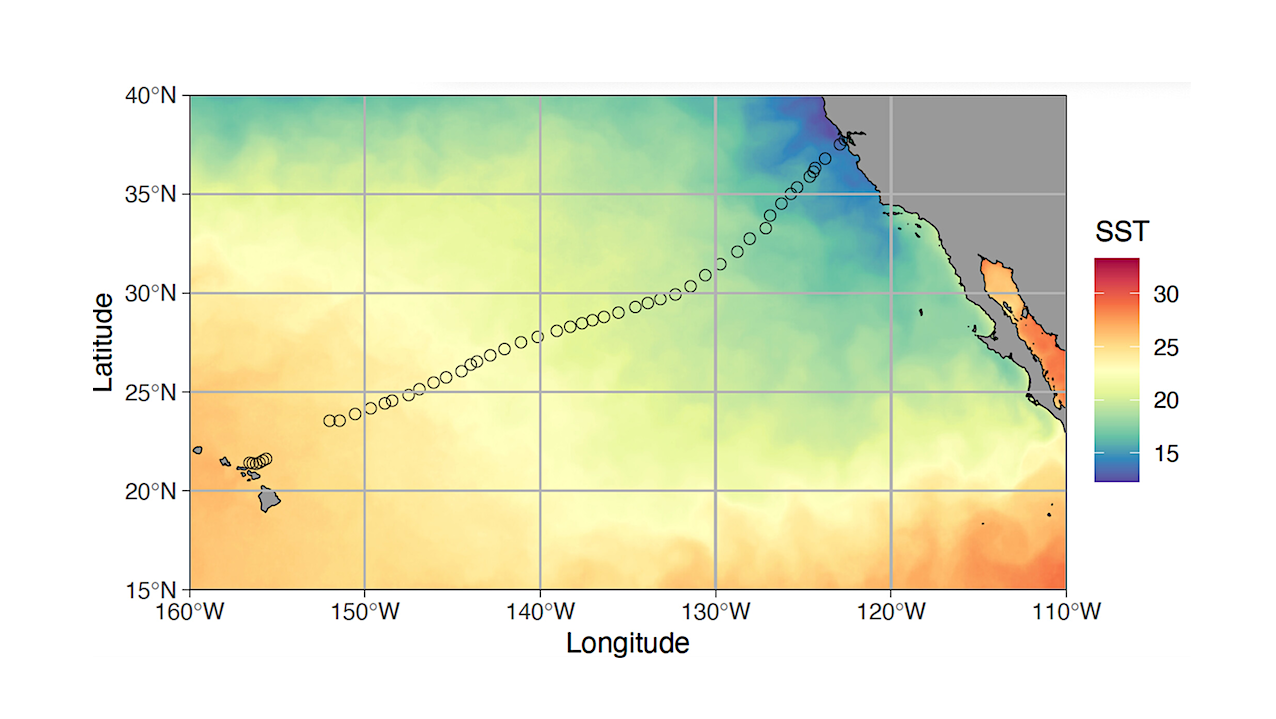
Here, we addressed those challenges by combining two robotic technologies: an uncrewed surface vessel (USV) fitted with an automated eDNA sample collection device to survey surface waters in the eastern North Pacific Ocean from Alameda, CA to Honolulu, HI. USV Surveyor SD 1200 (Saildrone) carrying the Environmental Sample Processor (ESP) collected 2-L water samples by filtration followed by RNA later preservation at regular intervals over a 4200-km, 29-day transit.
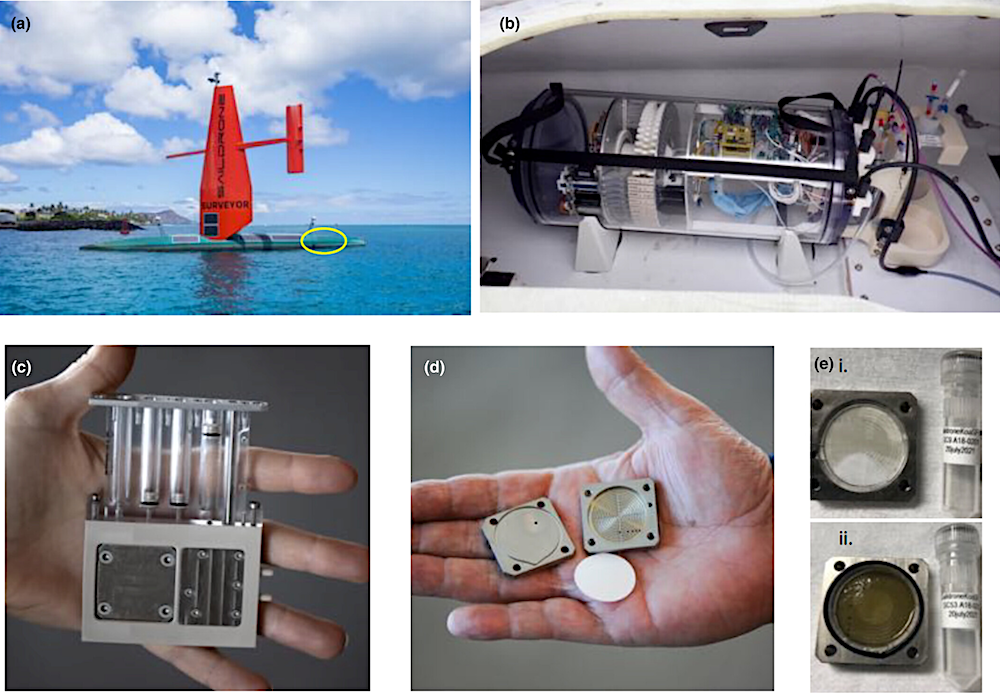
USV Surveyor SD 1200 reached Honolulu, HI after a 29-day cruise (a), carrying the Environmental Sample Processor in its aft hold (yellow circle). The ESP mounted in the hold (b). The ESP carried 60 cartridges where filtering and preservation of each eDNA sample took place (c). Each cartridge contained a square “puck” that housed a 25 mm, 0.2 μm filter (d). Opened pucks show biomass collected on filters from two eDNA preserved samples (e) collected off the coastlines of Hawaii (i) and California (ii).
Sixty samples (52 field and 8 controls) were acquired and used to estimate the concentration of specific genes and assess eukaryotic diversity via targeted qPCR and metabarcoding of the cytochrome oxidase subunit I (COI) gene, respectively. Comparisons of control samples revealed important considerations for interpreting results.
Samples stored at ambient temperatures onboard Surveyor over the length of the voyage had less total recoverable DNA and specific target gene concentrations compared to the same material immediately flash-frozen after collection and stored in a laboratory.
In contrast, the biodiversity of the COI genes in those samples was similar regardless of sample age and storage condition. COI genes affiliated with 40 eukaryotic phyla were found in native samples collected during the voyage.
The distribution and dominance of those phyla varied across different regions, with some taxa spanning large continuous stretches >2000 km, while others were only detected in a single sample.
This work highlights the utility and potential of using USVs fitted with autonomous eDNA sample collection devices to improve ocean exploration and support large, basin-scale, systematic biodiversity surveys.
Results of this study also inform future technical considerations for using automated eDNA samplers to acquire material and store it over prolonged periods under prevailing environmental conditions.
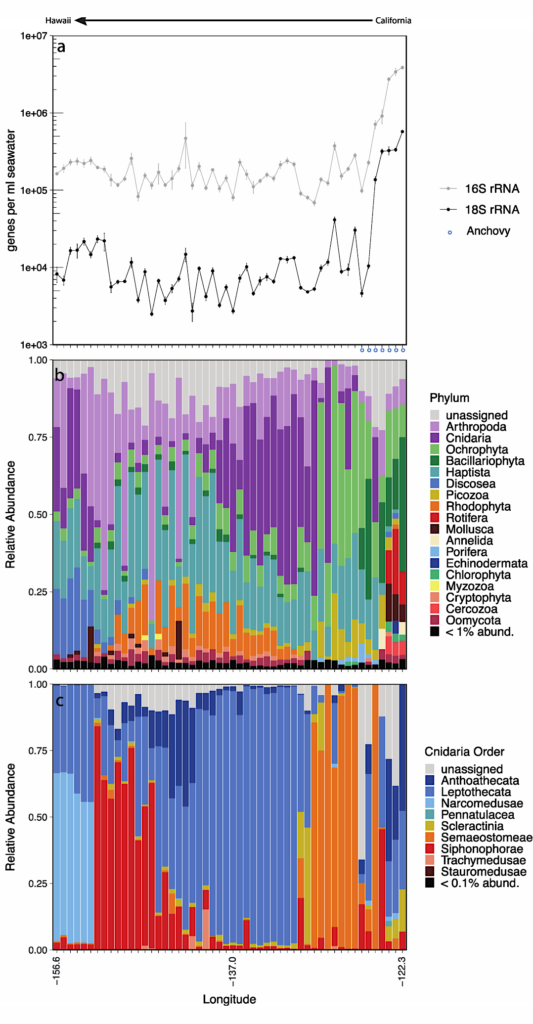
Analysis of eDNA from particulate samples collected and preserved by the ESP. The concentrations of targets as determined by qPCR (a) for 16S and 18S rRNA genes, and anchovy over the transit from California (right) to Hawaii (left). Anchovy dLoop genes were detected but not quantifiable in a subset of the samples nearest California (blue circles in panel a). The relative abundances of unrarified CO1 gene sequences affiliated with Eukaryotic phyla (56% of total) and Cnidarian orders are shown in panels b and c, respectively. CO1 gene sequences not identified as Eukarya (e.g., unknown, no_hit, Bacteria, and Archaea) were removed prior to analysis.
Autonomous eDNA collection using an uncrewed surface vessel over a 4200-km transect of the eastern Pacific Ocean, Wiley Online Library (Open access)
Astrobiology